[English] 日本語
 Yorodumi
Yorodumi- EMDB-25482: Cryo-EM structure of Arabidopsis Ago10-guide-target RNA complex i... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Arabidopsis Ago10-guide-target RNA complex in a bent duplex conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | miRNA / siRNA / RNA silencing / Argonaute / plant / GENE REGULATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of shoot apical meristem development / primary shoot apical meristem specification / regulation of meristem structural organization / miRNA metabolic process / regulatory ncRNA-mediated gene silencing / miRNA binding / plastid / somatic stem cell population maintenance / regulation of translation / defense response to virus ...regulation of shoot apical meristem development / primary shoot apical meristem specification / regulation of meristem structural organization / miRNA metabolic process / regulatory ncRNA-mediated gene silencing / miRNA binding / plastid / somatic stem cell population maintenance / regulation of translation / defense response to virus / ribonucleoprotein complex / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
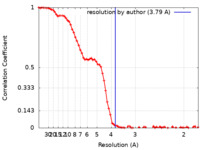
| Method | single particle reconstruction / cryo EM / Resolution: 3.79 Å | |||||||||
 Authors Authors | Xiao Y / MacRae IJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structural basis for RNA slicing by a plant Argonaute. Authors: Yao Xiao / Shintaro Maeda / Takanori Otomo / Ian J MacRae /  Abstract: Argonaute (AGO) proteins use small RNAs to recognize transcripts targeted for silencing in plants and animals. Many AGOs cleave target RNAs using an endoribonuclease activity termed 'slicing'. ...Argonaute (AGO) proteins use small RNAs to recognize transcripts targeted for silencing in plants and animals. Many AGOs cleave target RNAs using an endoribonuclease activity termed 'slicing'. Slicing by DNA-guided prokaryotic AGOs has been studied in detail, but structural insights into RNA-guided slicing by eukaryotic AGOs are lacking. Here we present cryogenic electron microscopy structures of the Arabidopsis thaliana Argonaute10 (AtAgo10)-guide RNA complex with and without a target RNA representing a slicing substrate. The AtAgo10-guide-target complex adopts slicing-competent and slicing-incompetent conformations that are unlike known prokaryotic AGO structures. AtAgo10 slicing activity is licensed by docking target (t) nucleotides t9-t13 into a surface channel containing the AGO endoribonuclease active site. A β-hairpin in the L1 domain secures the t9-t13 segment and coordinates t9-t13 docking with extended guide-target pairing. Results show that prokaryotic and eukaryotic AGOs use distinct mechanisms for achieving target slicing and provide insights into small interfering RNA potency. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25482.map.gz emd_25482.map.gz | 23.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25482-v30.xml emd-25482-v30.xml emd-25482.xml emd-25482.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25482_fsc.xml emd_25482_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_25482.png emd_25482.png | 51.9 KB | ||
| Filedesc metadata |  emd-25482.cif.gz emd-25482.cif.gz | 6.6 KB | ||
| Others |  emd_25482_additional_1.map.gz emd_25482_additional_1.map.gz emd_25482_half_map_1.map.gz emd_25482_half_map_1.map.gz emd_25482_half_map_2.map.gz emd_25482_half_map_2.map.gz | 20.9 MB 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25482 http://ftp.pdbj.org/pub/emdb/structures/EMD-25482 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25482 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25482 | HTTPS FTP |
-Validation report
| Summary document |  emd_25482_validation.pdf.gz emd_25482_validation.pdf.gz | 968 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25482_full_validation.pdf.gz emd_25482_full_validation.pdf.gz | 967.5 KB | Display | |
| Data in XML |  emd_25482_validation.xml.gz emd_25482_validation.xml.gz | 12.7 KB | Display | |
| Data in CIF |  emd_25482_validation.cif.gz emd_25482_validation.cif.gz | 17.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25482 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25482 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25482 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25482 | HTTPS FTP |
-Related structure data
| Related structure data |  7swqMC  7svaC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25482.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25482.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Local resolution filtered map
| File | emd_25482_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_25482_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_25482_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : AtAgo10-guide-target RNA complex
| Entire | Name: AtAgo10-guide-target RNA complex |
|---|---|
| Components |
|
-Supramolecule #1: AtAgo10-guide-target RNA complex
| Supramolecule | Name: AtAgo10-guide-target RNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: The complex of Arabidopsis Argonaute10 with a synthetic guide RNA and a target pairing to 2-16nt of guide guide |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Protein argonaute 10
| Macromolecule | Name: Protein argonaute 10 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 110.981633 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPIRQMKDSS ETHLVIKTQP LKHHNPKTVQ NGKIPPPSPS PVTVTTPATV TQSQASSPSP PSKNRSRRRN RGGRKSDQGD VCMRPSSRP RKPPPPSQTT SSAVSVATAG EIVAVNHQMQ MGVRKNSNFA PRPGFGTLGT KCIVKANHFL ADLPTKDLNQ Y DVTITPEV ...String: MPIRQMKDSS ETHLVIKTQP LKHHNPKTVQ NGKIPPPSPS PVTVTTPATV TQSQASSPSP PSKNRSRRRN RGGRKSDQGD VCMRPSSRP RKPPPPSQTT SSAVSVATAG EIVAVNHQMQ MGVRKNSNFA PRPGFGTLGT KCIVKANHFL ADLPTKDLNQ Y DVTITPEV SSKSVNRAII AELVRLYKES DLGRRLPAYD GRKSLYTAGE LPFTWKEFSV KIVDEDDGII NGPKRERSYK VA IKFVARA NMHHLGEFLA GKRADCPQEA VQILDIVLRE LSVKRFCPVG RSFFSPDIKT PQRLGEGLES WCGFYQSIRP TQM GLSLNI DMASAAFIEP LPVIEFVAQL LGKDVLSKPL SDSDRVKIKK GLRGVKVEVT HRANVRRKYR VAGLTTQPTR ELMF PVDEN CTMKSVIEYF QEMYGFTIQH THLPCLQVGN QKKASYLPME ACKIVEGQRY TKRLNEKQIT ALLKVTCQRP RDREN DILR TVQHNAYDQD PYAKEFGMNI SEKLASVEAR ILPAPWLKYH ENGKEKDCLP QVGQWNMMNK KMINGMTVSR WACVNF SRS VQENVARGFC NELGQMCEVS GMEFNPEPVI PIYSARPDQV EKALKHVYHT SMNKTKGKEL ELLLAILPDN NGSLYGD LK RICETELGLI SQCCLTKHVF KISKQYLANV SLKINVKMGG RNTVLVDAIS CRIPLVSDIP TIIFGADVTH PENGEESS P SIAAVVASQD WPEVTKYAGL VCAQAHRQEL IQDLYKTWQD PVRGTVSGGM IRDLLISFRK ATGQKPLRII FYRAGVSEG QFYQVLLYEL DAIRKACASL EPNYQPPVTF IVVQKRHHTR LFANNHRDKN STDRSGNILP GTVVDTKICH PTEFDFYLCS HAGIQGTSR PAHYHVLWDE NNFTADGIQS LTNNLCYTYA RCTRSVSIVP PAYYAHLAAF RARFYLEPEI MQDNGSPGKK N TKTTTVGD VGVKPLPALK ENVKRVMFYC UniProtKB: Protein argonaute 10 |
-Macromolecule #2: RNA (5'-R(P*UP*GP*GP*AP*GP*UP*GP*UP*GP*AP*CP*AP*AP*UP*GP*GP*UP*GP...
| Macromolecule | Name: RNA (5'-R(P*UP*GP*GP*AP*GP*UP*GP*UP*GP*AP*CP*AP*AP*UP*GP*GP*UP*GP*UP*UP*U)-3') type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 6.788021 KDa |
| Sequence | String: UGGAGUGUGA CAAUGGUGUU U |
-Macromolecule #3: RNA (5'-R(P*CP*CP*AP*UP*UP*GP*UP*CP*AP*CP*AP*CP*UP*CP*CP*AP*AP*A)-3')
| Macromolecule | Name: RNA (5'-R(P*CP*CP*AP*UP*UP*GP*UP*CP*AP*CP*AP*CP*UP*CP*CP*AP*AP*A)-3') type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 5.636419 KDa |
| Sequence | String: CCAUUGUCAC ACUCCAAA |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 2049 / Average electron dose: 66.88 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.7 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)