[English] 日本語
 Yorodumi
Yorodumi- EMDB-24783: CryoEM structure of Vibrio cholerae transposon Tn6677 AAA+ ATPase TnsC -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Vibrio cholerae transposon Tn6677 AAA+ ATPase TnsC | |||||||||
 Map data Map data | post-processed map with deepEMhacer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / transposon / ATPase / AAA+ / TnsC / DNA BINDING PROTEIN | |||||||||
| Biological species |  | |||||||||
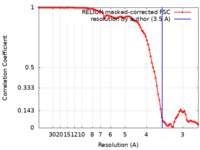
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Hoffmann FT / Kim M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Selective TnsC recruitment enhances the fidelity of RNA-guided transposition. Authors: Florian T Hoffmann / Minjoo Kim / Leslie Y Beh / Jing Wang / Phuc Leo H Vo / Diego R Gelsinger / Jerrin Thomas George / Christopher Acree / Jason T Mohabir / Israel S Fernández / Samuel H Sternberg /   Abstract: Bacterial transposons are pervasive mobile genetic elements that use distinct DNA-binding proteins for horizontal transmission. For example, Escherichia coli Tn7 homes to a specific attachment site ...Bacterial transposons are pervasive mobile genetic elements that use distinct DNA-binding proteins for horizontal transmission. For example, Escherichia coli Tn7 homes to a specific attachment site using TnsD, whereas CRISPR-associated transposons use type I or type V Cas effectors to insert downstream of target sites specified by guide RNAs. Despite this targeting diversity, transposition invariably requires TnsB, a DDE-family transposase that catalyses DNA excision and insertion, and TnsC, a AAA+ ATPase that is thought to communicate between transposase and targeting proteins. How TnsC mediates this communication and thereby regulates transposition fidelity has remained unclear. Here we use chromatin immunoprecipitation with sequencing to monitor in vivo formation of the type I-F RNA-guided transpososome, enabling us to resolve distinct protein recruitment events before integration. DNA targeting by the TniQ-Cascade complex is surprisingly promiscuous-hundreds of genomic off-target sites are sampled, but only a subset of those sites is licensed for TnsC and TnsB recruitment, revealing a crucial proofreading checkpoint. To advance the mechanistic understanding of interactions responsible for transpososome assembly, we determined structures of TnsC using cryogenic electron microscopy and found that ATP binding drives the formation of heptameric rings that thread DNA through the central pore, thereby positioning the substrate for downstream integration. Collectively, our results highlight the molecular specificity imparted by consecutive factor binding to genomic target sites during RNA-guided transposition, and provide a structural roadmap to guide future engineering efforts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24783.map.gz emd_24783.map.gz | 1.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24783-v30.xml emd-24783-v30.xml emd-24783.xml emd-24783.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24783_fsc.xml emd_24783_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_24783.png emd_24783.png | 112.4 KB | ||
| Masks |  emd_24783_msk_1.map emd_24783_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-24783.cif.gz emd-24783.cif.gz | 5.7 KB | ||
| Others |  emd_24783_additional_1.map.gz emd_24783_additional_1.map.gz emd_24783_additional_2.map.gz emd_24783_additional_2.map.gz emd_24783_half_map_1.map.gz emd_24783_half_map_1.map.gz emd_24783_half_map_2.map.gz emd_24783_half_map_2.map.gz | 49.7 MB 5.2 MB 49.7 MB 49.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24783 http://ftp.pdbj.org/pub/emdb/structures/EMD-24783 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24783 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24783 | HTTPS FTP |
-Validation report
| Summary document |  emd_24783_validation.pdf.gz emd_24783_validation.pdf.gz | 638.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24783_full_validation.pdf.gz emd_24783_full_validation.pdf.gz | 638.4 KB | Display | |
| Data in XML |  emd_24783_validation.xml.gz emd_24783_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  emd_24783_validation.cif.gz emd_24783_validation.cif.gz | 20.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24783 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24783 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24783 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24783 | HTTPS FTP |
-Related structure data
| Related structure data |  7rzyMC  7ufiC  7ufmC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24783.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24783.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post-processed map with deepEMhacer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36333 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_24783_msk_1.map emd_24783_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
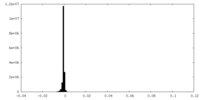
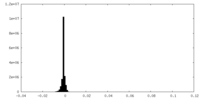
| Density Histograms |
-Additional map: unsharpened map
| File | emd_24783_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
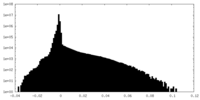
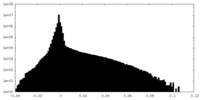
| Density Histograms |
-Additional map: Relion3 post-processed map
| File | emd_24783_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion3 post-processed map | ||||||||||||
| Projections & Slices |
| ||||||||||||
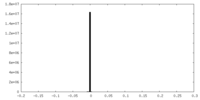
| Density Histograms |
-Half map: half-map-1
| File | emd_24783_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map-1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
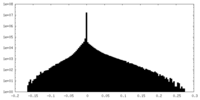
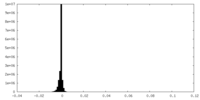
| Density Histograms |
-Half map: half-map-2
| File | emd_24783_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map-2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
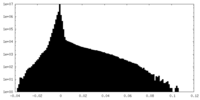
| Density Histograms |
- Sample components
Sample components
-Entire : VchTnsC
| Entire | Name: VchTnsC |
|---|---|
| Components |
|
-Supramolecule #1: VchTnsC
| Supramolecule | Name: VchTnsC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 238 KDa |
-Macromolecule #1: Tn6677 Vibrio cholerae transposon TnsC (VchTnsC)
| Macromolecule | Name: Tn6677 Vibrio cholerae transposon TnsC (VchTnsC) / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.596043 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSETREARIS RAKRAFVSTP SVRKILSYMD RCRDLSDLES EPTCMMVYGA SGVGKTTVIK KYLNQNRRES EAGGDIIPVL HIELPDNAK PVDAARELLV EMGDPLALYE TDLARLTKRL TELIPAVGVK LIIIDEFQHL VEERSNRVLT QVGNWLKMIL N KTKCPIVI ...String: MSETREARIS RAKRAFVSTP SVRKILSYMD RCRDLSDLES EPTCMMVYGA SGVGKTTVIK KYLNQNRRES EAGGDIIPVL HIELPDNAK PVDAARELLV EMGDPLALYE TDLARLTKRL TELIPAVGVK LIIIDEFQHL VEERSNRVLT QVGNWLKMIL N KTKCPIVI FGMPYSKVVL QANSQLHGRF SIQVELRPFS YNGGRGVFKT FLEYLDKALP FEKQAGLANE SLQKKLYAFS QG NMRSLRN LIYQASIEAI DNQHETITEE DFVFASKLTS GDKPNSWKNP FEEGVEVTED MLRPPPKDIG WEDYLRHSTP RVS KPGRNK NFFE |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 7 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Grid | Model: Homemade / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 80.0 K |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 51.0 µm / Calibrated defocus max: 1.9000000000000001 µm / Calibrated defocus min: 0.55 µm / Calibrated magnification: 65000 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 0.001 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 65000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: BACKBONE TRACE |
|---|---|
| Output model |  PDB-7rzy: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)