[English] 日本語
 Yorodumi
Yorodumi- EMDB-24570: Substance P bound to active human neurokinin 1 receptor in comple... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24570 | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Substance P bound to active human neurokinin 1 receptor in complex with miniGs399 | ||||||||||||||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||||||||
 Keywords Keywords | Substance P / G protein / GPCR / Neurokinin / Tachykinin / SIGNALING PROTEIN / SIGNALING PROTEIN-MEMBRANE PROTEIN complex | ||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsubstance P receptor binding / substance P receptor activity / positive regulation of corticosterone secretion / tachykinin receptor activity / positive regulation of flagellated sperm motility / aggressive behavior / Tachykinin receptors bind tachykinins / sperm ejaculation / positive regulation of uterine smooth muscle contraction / insemination ...substance P receptor binding / substance P receptor activity / positive regulation of corticosterone secretion / tachykinin receptor activity / positive regulation of flagellated sperm motility / aggressive behavior / Tachykinin receptors bind tachykinins / sperm ejaculation / positive regulation of uterine smooth muscle contraction / insemination / positive regulation of synaptic transmission, cholinergic / detection of abiotic stimulus / positive regulation of lymphocyte proliferation / tachykinin receptor signaling pathway / response to ozone / sperm head / operant conditioning / positive regulation of action potential / smooth muscle contraction involved in micturition / positive regulation of acute inflammatory response / response to auditory stimulus / positive regulation of blood pressure / positive regulation of vascular permeability / regulation of smooth muscle cell proliferation / positive regulation of hormone secretion / regulation of smooth muscle cell migration / positive regulation of ossification / positive regulation of leukocyte migration / eating behavior / negative regulation of heart rate / response to pain / behavioral response to pain / angiotensin-mediated drinking behavior / positive regulation of vasoconstriction / positive regulation of epithelial cell migration / response to electrical stimulus / PKA activation in glucagon signalling / associative learning / developmental growth / hair follicle placode formation / intracellular transport / D1 dopamine receptor binding / long-term memory / neuropeptide signaling pathway / neuronal dense core vesicle / sperm flagellum / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / positive regulation of stress fiber assembly / response to hormone / adenylate cyclase-activating adrenergic receptor signaling pathway / regulation of insulin secretion / sensory perception of pain / cellular response to glucagon stimulus / response to progesterone / adenylate cyclase activator activity / trans-Golgi network membrane / positive regulation of epithelial cell proliferation / positive regulation of synaptic transmission, GABAergic / response to nicotine / negative regulation of inflammatory response to antigenic stimulus / cellular response to nerve growth factor stimulus / bone development / regulation of blood pressure / platelet aggregation / cognition / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / sensory perception of smell / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / response to estradiol / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / sperm midpiece / adenylate cyclase-activating dopamine receptor signaling pathway Similarity search - Function | ||||||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | ||||||||||||||||||||||||||||||||||||
 Authors Authors | Harris JA / Faust B | ||||||||||||||||||||||||||||||||||||
| Funding support |  United States, United States,  Australia, 11 items Australia, 11 items
| ||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2022 Journal: Nat Chem Biol / Year: 2022Title: Selective G protein signaling driven by substance P-neurokinin receptor dynamics. Authors: Julian A Harris / Bryan Faust / Arisbel B Gondin / Marc André Dämgen / Carl-Mikael Suomivuori / Nicholas A Veldhuis / Yifan Cheng / Ron O Dror / David M Thal / Aashish Manglik /   Abstract: The neuropeptide substance P (SP) is important in pain and inflammation. SP activates the neurokinin-1 receptor (NK1R) to signal via G and G proteins. Neurokinin A also activates NK1R, but leads to ...The neuropeptide substance P (SP) is important in pain and inflammation. SP activates the neurokinin-1 receptor (NK1R) to signal via G and G proteins. Neurokinin A also activates NK1R, but leads to selective G signaling. How two stimuli yield distinct G protein signaling at the same G protein-coupled receptor remains unclear. We determined cryogenic-electron microscopy structures of active NK1R bound to SP or the G-biased peptide SP6-11. Peptide interactions deep within NK1R are critical for receptor activation. Conversely, interactions between SP and NK1R extracellular loops are required for potent G signaling but not G signaling. Molecular dynamics simulations showed that these superficial contacts restrict SP flexibility. SP6-11, which lacks these interactions, is dynamic while bound to NK1R. Structural dynamics of NK1R agonists therefore depend on interactions with the receptor extracellular loops and regulate G protein signaling selectivity. Similar interactions between other neuropeptides and their cognate receptors may tune intracellular signaling. | ||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24570.map.gz emd_24570.map.gz | 115.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24570-v30.xml emd-24570-v30.xml emd-24570.xml emd-24570.xml | 28.9 KB 28.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24570.png emd_24570.png | 50.3 KB | ||
| Filedesc metadata |  emd-24570.cif.gz emd-24570.cif.gz | 7.4 KB | ||
| Others |  emd_24570_additional_1.map.gz emd_24570_additional_1.map.gz emd_24570_half_map_1.map.gz emd_24570_half_map_1.map.gz emd_24570_half_map_2.map.gz emd_24570_half_map_2.map.gz | 116.1 MB 16.8 MB 16.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24570 http://ftp.pdbj.org/pub/emdb/structures/EMD-24570 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24570 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24570 | HTTPS FTP |
-Related structure data
| Related structure data |  7rmhMC  7rmgC  7rmiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10786 (Title: Cryo electron microscopy final particle stacks of Substance P-Neurokinin Receptor G protein complexes EMPIAR-10786 (Title: Cryo electron microscopy final particle stacks of Substance P-Neurokinin Receptor G protein complexesData size: 170.8 Data #1: Particle stack and final .star file for SP-NK1R-miniGs399 reconstruction [picked particles - single frame - processed] Data #2: Particle stack and final .star file for SP6-11-NK1R-miniGsq70 reconstruction [picked particles - single frame - processed] Data #3: Particle stack and final .star file for SP-NK1R-miniGsq70 reconstruction [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24570.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24570.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
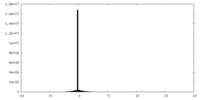
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_24570_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_24570_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_24570_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Substance P bound to Neurokinin1 receptor-miniGs399 complex
+Supramolecule #1: Substance P bound to Neurokinin1 receptor-miniGs399 complex
+Supramolecule #2: Substance P
+Supramolecule #3: Substance-P receptor
+Supramolecule #4: miniGs399
+Supramolecule #5: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1/...
+Supramolecule #6: Nanobody 35
+Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
+Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
+Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
+Macromolecule #4: Nanobody 35
+Macromolecule #5: Substance-P receptor
+Macromolecule #6: Substance P
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 3670 / Average exposure time: 1.52 sec. / Average electron dose: 49.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)












































 Trichoplusia ni (cabbage looper)
Trichoplusia ni (cabbage looper)

