+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24238 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Melbournevirus nucleosome like particle | |||||||||
 Map data Map data | Melbournevirus nucleosome like particle | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nucleosome / Virus / DNA BINDING PROTEIN / DNA BINDING PROTEIN-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationchromosome condensation / virion component / structural constituent of chromatin / host cell cytoplasm / protein heterodimerization activity / host cell nucleus / DNA binding Similarity search - Function | |||||||||
| Biological species |  Melbournevirus / Melbournevirus /  | |||||||||
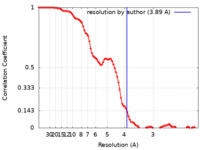
| Method | single particle reconstruction / cryo EM / Resolution: 3.89 Å | |||||||||
 Authors Authors | Liu Y / Toner CM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: Virus-encoded histone doublets are essential and form nucleosome-like structures. Authors: Yang Liu / Hugo Bisio / Chelsea Marie Toner / Sandra Jeudy / Nadege Philippe / Keda Zhou / Samuel Bowerman / Alison White / Garrett Edwards / Chantal Abergel / Karolin Luger /   Abstract: The organization of genomic DNA into defined nucleosomes has long been viewed as a hallmark of eukaryotes. This paradigm has been challenged by the identification of "minimalist" histones in archaea ...The organization of genomic DNA into defined nucleosomes has long been viewed as a hallmark of eukaryotes. This paradigm has been challenged by the identification of "minimalist" histones in archaea and more recently by the discovery of genes that encode fused remote homologs of the four eukaryotic histones in Marseilleviridae, a subfamily of giant viruses that infect amoebae. We demonstrate that viral doublet histones are essential for viral infectivity, localize to cytoplasmic viral factories after virus infection, and ultimately are found in the mature virions. Cryogenic electron microscopy (cryo-EM) structures of viral nucleosome-like particles show strong similarities to eukaryotic nucleosomes despite the limited sequence identify. The unique connectors that link the histone chains contribute to the observed instability of viral nucleosomes, and some histone tails assume structural roles. Our results further expand the range of "organisms" that require nucleosomes and suggest a specialized function of histones in the biology of these unusual viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24238.map.gz emd_24238.map.gz | 2.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24238-v30.xml emd-24238-v30.xml emd-24238.xml emd-24238.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24238_fsc.xml emd_24238_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_24238.png emd_24238.png | 22.3 KB | ||
| Masks |  emd_24238_msk_1.map emd_24238_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-24238.cif.gz emd-24238.cif.gz | 5.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24238 http://ftp.pdbj.org/pub/emdb/structures/EMD-24238 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24238 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24238 | HTTPS FTP |
-Validation report
| Summary document |  emd_24238_validation.pdf.gz emd_24238_validation.pdf.gz | 393.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24238_full_validation.pdf.gz emd_24238_full_validation.pdf.gz | 392.8 KB | Display | |
| Data in XML |  emd_24238_validation.xml.gz emd_24238_validation.xml.gz | 10.9 KB | Display | |
| Data in CIF |  emd_24238_validation.cif.gz emd_24238_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24238 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24238 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24238 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24238 | HTTPS FTP |
-Related structure data
| Related structure data |  7n8nMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24238.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24238.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Melbournevirus nucleosome like particle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
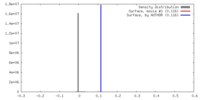
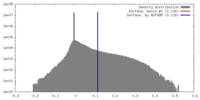
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_24238_msk_1.map emd_24238_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
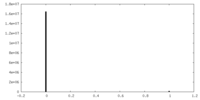
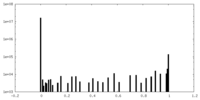
| Density Histograms |
- Sample components
Sample components
-Entire : Melbournevirus nucleosome like particle
| Entire | Name: Melbournevirus nucleosome like particle |
|---|---|
| Components |
|
-Supramolecule #1: Melbournevirus nucleosome like particle
| Supramolecule | Name: Melbournevirus nucleosome like particle / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 218 KDa |
-Supramolecule #2: Histone H4-H3 doublet, Histone H2B-H2A doublet
| Supramolecule | Name: Histone H4-H3 doublet, Histone H2B-H2A doublet / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Melbournevirus Melbournevirus |
-Supramolecule #3: DNA 147-mer
| Supramolecule | Name: DNA 147-mer / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Histone H4-H3 doublet
| Macromolecule | Name: Histone H4-H3 doublet / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Melbournevirus Melbournevirus |
| Molecular weight | Theoretical: 26.73718 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGKP IPNPLLGLDS TENLYFQGSS KAGKKVKAQQ HGHLADHVSV GETQIPKAST QHLLRKAGSL SAAGDTEVPI RGFVHMKLH KLVQKSLLAM QLAKRKTIMK SDVKKAAELM HLPVFAIPTK DSGAKGSVFL SCRQKGAGSA GTGSETNSQE V RSQMKSTC ...String: MHHHHHHGKP IPNPLLGLDS TENLYFQGSS KAGKKVKAQQ HGHLADHVSV GETQIPKAST QHLLRKAGSL SAAGDTEVPI RGFVHMKLH KLVQKSLLAM QLAKRKTIMK SDVKKAAELM HLPVFAIPTK DSGAKGSVFL SCRQKGAGSA GTGSETNSQE V RSQMKSTC LIIPKERFRT MAKEISKKEG HDVHIAEAAL DMLQVIVESC TVRLLEKALV ITYSGKRTRV TSKDIETAFM LE HGPL UniProtKB: Histone doublet H4-H3 |
-Macromolecule #2: Histone H2B-H2A doublet
| Macromolecule | Name: Histone H2B-H2A doublet / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Melbournevirus Melbournevirus |
| Molecular weight | Theoretical: 32.05816 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGKP IPNPLLGLDS TENLYFQGSA TQKETTRKRD KSVNFRLGLR NMLAQIHPDI SVQTEALSEL SNIAVFLGKK ISHGAVTLL PEGTKTIKSS AVLLAAGDLY GKDLGRHAVG EMTKAVTRYG SAKESKEGSR SSKAKLQISV ARSERLLREH G GCSRVSEG ...String: MHHHHHHGKP IPNPLLGLDS TENLYFQGSA TQKETTRKRD KSVNFRLGLR NMLAQIHPDI SVQTEALSEL SNIAVFLGKK ISHGAVTLL PEGTKTIKSS AVLLAAGDLY GKDLGRHAVG EMTKAVTRYG SAKESKEGSR SSKAKLQISV ARSERLLREH G GCSRVSEG AAVALAAAIE YFMGEVLELA GNAARDSKKV RISVKHITLA IQNDAALFAV VGKGVFSGAG VSLISVPIPR KK ARKTTEK EASSPKKKAA PKKKKAASKQ KKSLSDKELA KLTKKELAKY EKEQGMSPGY UniProtKB: Histone doublet H2B-H2A |
-Macromolecule #3: DNA (147-MER)
| Macromolecule | Name: DNA (147-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.153781 KDa |
| Sequence | String: (DA)(DT)(DC)(DT)(DG)(DA)(DG)(DA)(DA)(DT) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG) (DA)(DC)(DA)(DG)(DC)(DT) ...String: (DA)(DT)(DC)(DT)(DG)(DA)(DG)(DA)(DA)(DT) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG) (DA)(DC)(DA)(DG)(DC)(DT)(DC)(DT) (DA)(DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT) (DA)(DA) (DA)(DC)(DG)(DC)(DA)(DC)(DG) (DT)(DA)(DC)(DG)(DC)(DG)(DC)(DT)(DG)(DT) (DC)(DC)(DC) (DC)(DC)(DG)(DC)(DG)(DT) (DT)(DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC) (DA)(DA)(DG)(DG) (DG)(DG)(DA)(DT)(DT) (DA)(DC)(DT)(DC)(DC)(DC)(DT)(DA)(DG)(DT) (DC)(DT)(DC)(DC)(DA) (DG)(DG)(DC)(DA) (DC)(DG)(DT)(DG)(DT)(DC)(DA)(DG)(DA)(DT) (DA)(DT)(DA)(DT)(DA)(DC) (DA)(DT)(DC) (DC)(DG)(DA)(DT) |
-Macromolecule #4: DNA (147-MER)
| Macromolecule | Name: DNA (147-MER) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.594043 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC) ...String: (DA)(DT)(DC)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC)(DC)(DC) (DC)(DT)(DT)(DG)(DG)(DC)(DG)(DG)(DT)(DT) (DA)(DA) (DA)(DA)(DC)(DG)(DC)(DG)(DG) (DG)(DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC) (DG)(DT)(DA) (DC)(DG)(DT)(DG)(DC)(DG) (DT)(DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT) (DG)(DC)(DT)(DA) (DG)(DA)(DG)(DC)(DT) (DG)(DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC) (DA)(DA)(DT)(DT)(DG) (DA)(DG)(DC)(DG) (DG)(DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC) (DC)(DG)(DG)(DA)(DT)(DT) (DC)(DT)(DC) (DA)(DG)(DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 400 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)