+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24181 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of bovine thyroglobulin with iodinated tyrosines | |||||||||
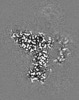
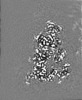
 Map data Map data | Combined EM map of reconstructions from focused refinements around the region surrounding half of the dimeric Tg molecule and the arm region of a Tg monomer - resampled in finer grid points. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | thyroid hormone synthesis / HORMONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationhormone biosynthetic process / thyroid hormone generation / hormone activity / extracellular space Similarity search - Function | |||||||||
| Biological species |  | |||||||||
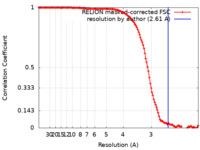
| Method | single particle reconstruction / cryo EM / Resolution: 2.61 Å | |||||||||
 Authors Authors | Kim K / Clarke OB | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2021 Journal: Acta Crystallogr D Struct Biol / Year: 2021Title: The structure of natively iodinated bovine thyroglobulin. Authors: Kookjoo Kim / Mykhailo Kopylov / Daija Bobe / Kotaro Kelley / Edward T Eng / Peter Arvan / Oliver B Clarke /  Abstract: Thyroglobulin is a homodimeric glycoprotein that is essential for the generation of thyroid hormones in vertebrates. Upon secretion into the lumen of follicles in the thyroid gland, tyrosine residues ...Thyroglobulin is a homodimeric glycoprotein that is essential for the generation of thyroid hormones in vertebrates. Upon secretion into the lumen of follicles in the thyroid gland, tyrosine residues within the protein become iodinated to produce monoiodotyrosine (MIT) and diiodotyrosine (DIT). A subset of evolutionarily conserved pairs of DIT (and MIT) residues can then engage in oxidative coupling reactions that yield either thyroxine (T; produced from coupling of a DIT `acceptor' with a DIT `donor') or triiodothyronine (T; produced from coupling of a DIT acceptor with an MIT donor). Although multiple iodotyrosine residues have been identified as potential donors and acceptors, the specificity and structural context of the pairings (i.e. which donor is paired with which acceptor) have remained unclear. Here, single-particle cryogenic electron microscopy (cryoEM) was used to generate a high-resolution reconstruction of bovine thyroglobulin (2.3 Å resolution in the core region and 2.6 Å overall), allowing the structural characterization of two post-reaction acceptor-donor pairs as well as tyrosine residues modified as MIT and DIT. A substantial spatial separation between donor Tyr149 and acceptor Tyr24 was observed, suggesting that for thyroxine synthesis significant peptide motion is required for coupling at the evolutionarily conserved thyroglobulin amino-terminus. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24181.map.gz emd_24181.map.gz | 459.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24181-v30.xml emd-24181-v30.xml emd-24181.xml emd-24181.xml | 45 KB 45 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24181_fsc.xml emd_24181_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_24181.png emd_24181.png | 86.8 KB | ||
| Masks |  emd_24181_msk_1.map emd_24181_msk_1.map emd_24181_msk_2.map emd_24181_msk_2.map | 178 MB 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-24181.cif.gz emd-24181.cif.gz | 9.3 KB | ||
| Others |  emd_24181_additional_1.map.gz emd_24181_additional_1.map.gz emd_24181_additional_10.map.gz emd_24181_additional_10.map.gz emd_24181_additional_2.map.gz emd_24181_additional_2.map.gz emd_24181_additional_3.map.gz emd_24181_additional_3.map.gz emd_24181_additional_4.map.gz emd_24181_additional_4.map.gz emd_24181_additional_5.map.gz emd_24181_additional_5.map.gz emd_24181_additional_6.map.gz emd_24181_additional_6.map.gz emd_24181_additional_7.map.gz emd_24181_additional_7.map.gz emd_24181_additional_8.map.gz emd_24181_additional_8.map.gz emd_24181_additional_9.map.gz emd_24181_additional_9.map.gz emd_24181_half_map_1.map.gz emd_24181_half_map_1.map.gz emd_24181_half_map_2.map.gz emd_24181_half_map_2.map.gz | 139.7 MB 10.3 MB 89.5 MB 137.5 MB 139.6 MB 139.6 MB 140.9 MB 140.7 MB 21.2 MB 74.4 MB 139 MB 139 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24181 http://ftp.pdbj.org/pub/emdb/structures/EMD-24181 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24181 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24181 | HTTPS FTP |
-Validation report
| Summary document |  emd_24181_validation.pdf.gz emd_24181_validation.pdf.gz | 911.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24181_full_validation.pdf.gz emd_24181_full_validation.pdf.gz | 911.3 KB | Display | |
| Data in XML |  emd_24181_validation.xml.gz emd_24181_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  emd_24181_validation.cif.gz emd_24181_validation.cif.gz | 27.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24181 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24181 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24181 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24181 | HTTPS FTP |
-Related structure data
| Related structure data |  7n4yMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10833 (Title: The structure of natively iodinated bovine thyroglobulin EMPIAR-10833 (Title: The structure of natively iodinated bovine thyroglobulinData size: 1.2 TB Data #1: Unaligned multi-frame micrographs of bovine thyroglobulin [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24181.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24181.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Combined EM map of reconstructions from focused refinements around the region surrounding half of the dimeric Tg molecule and the arm region of a Tg monomer - resampled in finer grid points. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.7453 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
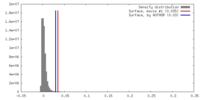
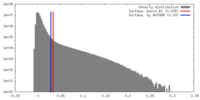
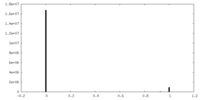
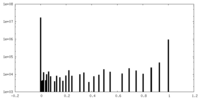
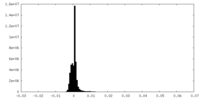
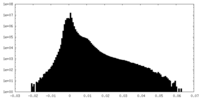
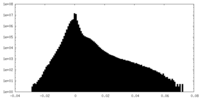
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Mask #1
+Mask #2
+Additional map: EM map of the focused refinement on the arm region of a Tg monomer
+Additional map: Combined EM map of density-modified maps of focused...
+Additional map: EM map of the global consensus refinement -...
+Additional map: EM map of the focused refinement on the...
+Additional map: The second EM half map of the focused...
+Additional map: The first EM half map of the focused...
+Additional map: The second EM half map of the focused...
+Additional map: The first EM half map of the focused...
+Additional map: density-modified EM map of the focused refinement on...
+Additional map: density-modified EM map of the focused refinement on...
+Half map: The first EM half map of the global consensus reconstruction
+Half map: The second EM half map of the global consensus reconstruction
- Sample components
Sample components
-Entire : Bovine thyroglobulin purified from bovine thyroid
| Entire | Name: Bovine thyroglobulin purified from bovine thyroid |
|---|---|
| Components |
|
-Supramolecule #1: Bovine thyroglobulin purified from bovine thyroid
| Supramolecule | Name: Bovine thyroglobulin purified from bovine thyroid / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 660 KDa |
-Macromolecule #1: Thyroglobulin
| Macromolecule | Name: Thyroglobulin / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 303.087844 KDa |
| Sequence | String: NIFE(T44)QVDAQ PLRPCELQRE RAFLKREDYV PQCAEDGSFQ TVQCGKDGAS CWCVDADGRE VPGSRQPGRP AACLSF CQL QKQQILLSSY INSTATSYLP QCQDSGDYSP VQCDLRRRQC WCVDAEGMEV YGTRQQGRPA RCPRSCEIRN RRLLHGV GD RSPPQCSPDG ...String: NIFE(T44)QVDAQ PLRPCELQRE RAFLKREDYV PQCAEDGSFQ TVQCGKDGAS CWCVDADGRE VPGSRQPGRP AACLSF CQL QKQQILLSSY INSTATSYLP QCQDSGDYSP VQCDLRRRQC WCVDAEGMEV YGTRQQGRPA RCPRSCEIRN RRLLHGV GD RSPPQCSPDG AFRPVQCKFV NTTDMMIFDL VHSYSRFPDA FVTFSSFRSR FPEVSGYCYC ADSQGRELAE TGLELLLD E IYDTIFAGLD LASTFAETTL YRILQRRFLA VQLVISGRFR CPTKCEVERF AATSFRHPYV PSCHPDGEYQ AAQCQQGGP CWCVDSRGQE IPGTRQRGEP PSCAEDQSCP SERRRAFSRL RFGPSGYFSR RSLLLAPEEG PVSQRFARFT ASCPPSIKEL FLDSGIFQP MLQGRDTRFV APESLLKEAI RGLFPSRELA RLALQFTTNA KRLQQNLFGG RFLVNVGQFN LSGALGTRGT F NFSHFFQQ LGLPGFQDGR ALADLAKPLS VGLNSNPASE APKASKIDVA LRKPVVGSFG FEVNLQENQN ALQFLSSFLE LP EFLLFLQ HAISVPEDIA RDLGDVMEMV FSSQGCGQAP GSLFVPACTA EGSYEEVQCF AGDCWCVDAQ GRELAGSRVR GGR PRCPTE CEKQRARMQS LLGSQPAGSS LFVPACTSKG NFLPVQCFNS ECYCVDTEGQ PIPGTRSALG EPKKCPSPCQ LQAE RAFLG TVRTLVSNPS TLPALSSIYI PQCSASGQWS PVQCDGPPEQ AFEWYERWEA QNSAGQALTP AELLMKIMSY REAAS RNFR LFIQNLYEAG QQGIFPGLAR YSSFQDVPVS VLEGNQTQPG GNVFLEPYLF WQILNGQLDR YPGPYSDFSA PLAHFD LRS CWCVDEAGQK LEGTRNEPNK VPACPGSCEE VKLRVLQFIR EAEEIVTYSN SSRFPLGESF LAAKGIRLTD EELAFPP LS PSRETFLEKF LSGSDYAIRL AAQSTFDFYQ RRLVTLAESP RAPSPVWSSA YLPQCDAFGG WEPVQCHAAT GHCWCVDG K GEYVPTSLTA RSRQIPQCPT SCERLRASGL LSSWKQAGVQ AEPSPKDLFI PTCLETGEFA RLQASEAGTW CVDPASGEG VPPGTNSSAQ CPSLCEVLQS GVPSRRTSPG YSPACRAEDG GFSPVQCDPA QGSCWCVLGS GEEVPGTRVA GSQPACESPQ CPLPFSVAD VAGGAILCER ASGLGAAAGQ RCQLRCSQGY RSAFPPEPLL CSVQRRRWES RPPQPRACQR PQFWQTLQTQ A QFQLLLPL GKVCSADYSG LLLAFQVFLL DELTARGFCQ IQVKTAGTPV SIPVCDDSSV KVECLSRERL GVNITWKLQL VD APPASLP DLQDVEEALA GKYLAGRFAD LIQSGTFQLH LDSKTFSADT SIRFLQGDRF GISPRTQFGC LEGFGRVVAA SDA SQDALG CVKCPEGSYF QDEQCIPCPA GFYQEQAGSL ACVPCPEGRT TVYAGAFSQT HCVTDCQKNE VGLQCDQDGQ YRAS QRDRT SGKAFCVDGE GRRLPWTEAE APLVDAQCLV MRKFEKLPES KVIFSADVAV MVRSEVPGSE SSLMQCLADC ALDEA CGFL TVSTAGSEVS CDFYAWASDS IACTTSGRSE DALGTSQATS FGSLQCQVKV RSREGDPLAV YLKKGQEFTI TGQKRF EQT GFQSALSGMY SPVTFSASGA SLAEVHLFCL LACDHDSCCD GFILVQVQGG PLLCGLLSSP DVLLCHVRDW RDPAEAQ AN ASCPGVTYDQ DSRQVTLRLG GQEIRGLTPL EGTQDTLTSF QQVYLWKDSD MGSRSESMGC RRDTEPRPAS PSETDLTT G LFSPVDLIQV IVDGNVSLPS QQHWLFKHLF SLQQANLWCL SRCAGEPSFC QLAEVTDSEP LYFTCTLYPE AQVCDDILE SSPKGCRLIL PRRPSALYRK KVVLQDRVKN FYNRLPFQKL TGISIRNKVP MSDKSISSGF FECERLCDMD PCCTGFGFLN VSQLKGGEV TCLTLNSLGL QTCSEE(TYI)GGV WRILDCGSPD TEVRTYPFGW YQKPVSPSDA PSFCPSVALP ALTENVA LD SWQSLALSSV IVDPSIRNFD VAHISTAAVG NFSAARDRCL WECSRHQDCL VTTLQTQPGA VRCMFYADTQ SCTHSLQA Q NCRLLLHEEA TYIYRKPNIP LPGFGTSSPS VPIATHGQLL GRSQAIQVGT SWKPVDQFLG VPYAAPPLGE KRFRAPEHL NWTGSWEATK PRARCWQPGI RTPTPPGVSE DCLYLNVFVP QNMPPNASVL VFFHNAAEGK GSGDRPAVDG SFLAAVGNLI VVTASYRTG IFGFLSSGSS ELSGNWGLLD QVVALTWVQT HIQAFGGDPR RVTLAADRGG ADIASIHLVT TRAANSRLFR R AVLMGGSA LSPAAVIRPE RARQQAAALA KEVGCPSSSV QEMVSCLRQE PARILNDAQT KLLAVSGPFH YWGPVVDGQY LR ETPARVL QRAPRVKVDL LIGSSQDDGL INRAKAVKQF EESQGRTSSK TAFYQALQNS LGGEAADAGV QAAATWYYSL EHD SDD(T44)AS FSRALEQATR DYFIICPVID MASHWARTVR GNVFMYHAPE SYSHSSLELL TDVLYAFGLP FYPAYEGQFT LEEKSLSLK IMQYFSNFIR SGNPNYPHEF SRRAPEFAAP WPDFVPRDGA ESYKELSVLL PNRQGLKKAD CSFWSKYIQS L KASADETK DGPSADSEEE DQPAGSGLTE DLLGLPELAS KTYSK UniProtKB: Thyroglobulin |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 10 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 308 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 71.32 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)