[English] 日本語
 Yorodumi
Yorodumi- EMDB-23997: Structure of the mammalian Transport Protein Particle Complex (TR... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23997 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the mammalian Transport Protein Particle Complex (TRAPP) III | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationconstitutive secretory pathway / regulation of kinetochore assembly / vesicle coating / vesicle tethering / positive regulation of protein localization to kinetochore / TRAPPII protein complex / TRAPPIII protein complex / TRAPP complex / collagen biosynthetic process / metaphase chromosome alignment ...constitutive secretory pathway / regulation of kinetochore assembly / vesicle coating / vesicle tethering / positive regulation of protein localization to kinetochore / TRAPPII protein complex / TRAPPIII protein complex / TRAPP complex / collagen biosynthetic process / metaphase chromosome alignment / COPII vesicle coating / regulation of protein complex stability / RAB GEFs exchange GTP for GDP on RABs / Golgi organization / endoplasmic reticulum-Golgi intermediate compartment / endoplasmic reticulum to Golgi vesicle-mediated transport / kinetochore / Golgi apparatus / nucleoplasm / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
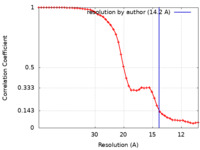
| Method | single particle reconstruction / negative staining / Resolution: 14.2 Å | |||||||||||||||
 Authors Authors | Harris NJ / Dalwadi U / Yip CK / Burke JE / Jenkins ML | |||||||||||||||
| Funding support |  Canada, 4 items Canada, 4 items
| |||||||||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2021 Journal: J Mol Biol / Year: 2021Title: Biochemical Insight into Novel Rab-GEF Activity of the Mammalian TRAPPIII Complex. Authors: Noah J Harris / Meredith L Jenkins / Udit Dalwadi / Kaelin D Fleming / Sung-Eun Nam / Matthew A H Parson / Calvin K Yip / John E Burke /  Abstract: Transport Protein Particle complexes (TRAPP) are evolutionarily conserved regulators of membrane trafficking, with this mediated by their guanine nucleotide exchange factor (GEF) activity towards Rab ...Transport Protein Particle complexes (TRAPP) are evolutionarily conserved regulators of membrane trafficking, with this mediated by their guanine nucleotide exchange factor (GEF) activity towards Rab GTPases. In metazoans evidence suggests that two different TRAPP complexes exist, TRAPPII and TRAPPIII. These two complexes share a common core of subunits, with complex specific subunits (TRAPPC9 and TRAPPC10 in TRAPPII and TRAPPC8, TRAPPC11, TRAPPC12, TRAPPC13 in TRAPPIII). TRAPPII and TRAPPIII have distinct specificity for GEF activity towards Rabs, with TRAPPIII acting on Rab1, and TRAPPII acting on Rab1 and Rab11. The molecular basis for how these complex specific subunits alter GEF activity towards Rab GTPases is unknown. Here we have used a combination of biochemical assays, hydrogen deuterium exchange mass spectrometry (HDX-MS) and electron microscopy to examine the regulation of TRAPPII and TRAPPIIII complexes in solution and on membranes. GEF assays revealed that TRAPPIII has GEF activity against Rab1 and Rab43, with no detectable activity against the other 18 Rabs tested. The TRAPPIII complex had significant differences in protein dynamics at the Rab binding site compared to TRAPPII, potentially indicating an important role of accessory subunits in altering the active site of TRAPP complexes. Both the TRAPPII and TRAPPIII complexes had enhanced GEF activity on lipid membranes, with HDX-MS revealing numerous conformational changes that accompany membrane association. HDX-MS also identified a membrane binding site in TRAPPC8. Collectively, our results provide insight into the functions of TRAPP complexes and how they can achieve Rab specificity. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23997.map.gz emd_23997.map.gz | 50.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23997-v30.xml emd-23997-v30.xml emd-23997.xml emd-23997.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23997_fsc.xml emd_23997_fsc.xml | 4.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_23997.png emd_23997.png | 68.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23997 http://ftp.pdbj.org/pub/emdb/structures/EMD-23997 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23997 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23997 | HTTPS FTP |
-Validation report
| Summary document |  emd_23997_validation.pdf.gz emd_23997_validation.pdf.gz | 324.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23997_full_validation.pdf.gz emd_23997_full_validation.pdf.gz | 324.1 KB | Display | |
| Data in XML |  emd_23997_validation.xml.gz emd_23997_validation.xml.gz | 9.5 KB | Display | |
| Data in CIF |  emd_23997_validation.cif.gz emd_23997_validation.cif.gz | 11.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23997 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23997 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23997 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23997 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23997.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23997.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.27 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
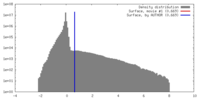
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Transport Protein Particle Complex III (TRAPP III)
| Entire | Name: Transport Protein Particle Complex III (TRAPP III) |
|---|---|
| Components |
|
-Supramolecule #1: Transport Protein Particle Complex III (TRAPP III)
| Supramolecule | Name: Transport Protein Particle Complex III (TRAPP III) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: Mammalian TRAPP III purified from Sf9 insect cells |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Freshly prepared gel filtration buffer, filtered through 0.22um filter and degassed | ||||||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Formate Details: Negative stained EM samples prepared by adsorbing sample on grid for 15s, followed by blotting of sample, 2 washes with water, 1 wash with stain and a final 30s soak in stain. | ||||||||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa Details: Glow discharged using a Pelco EasiGlow. 15mA current. | ||||||||||||
| Details | TRAPP III Complex was purified to homogeneity using affinity chromatography followed by size exclusion chromatography. Samples were kept at 4 degrees and shipped on wet ice for single particle EM analysis. |
- Electron microscopy
Electron microscopy
| Microscope | TFS TALOS L120C |
|---|---|
| Image recording | Film or detector model: FEI CETA (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 2.27 µm / Number grids imaged: 1 / Number real images: 100 / Average exposure time: 1.0 sec. / Average electron dose: 5.0 e/Å2 Details: Images were manually collected every 200nm on a grid square after a 30s delay for stage stabilization. Focus was calibrated every 10 images using thin rings in the power spectra. |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus min: 1.2 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos L120C / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)