[English] 日本語
 Yorodumi
Yorodumi- EMDB-23825: Glutamate synthase, glutamate dehydrogenase counter-enzyme comple... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23825 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Glutamate synthase, glutamate dehydrogenase counter-enzyme complex (GudB6-GltA6-GltB6) | |||||||||
 Map data Map data | Single GudB-GltAB asymmetric unit from GudB6-GltA6B6 complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Counter-enzyme complex / glutamate synthase / glutamate dehydrogenae / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationglutamate synthase activity / oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor / glutamate biosynthetic process / glutamate dehydrogenase (NAD+) activity / amino acid metabolic process / 3 iron, 4 sulfur cluster binding / glutamine metabolic process / iron-sulfur cluster binding / nucleotide binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
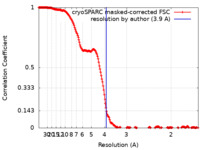
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Jayaraman V / Lee DJ | |||||||||
| Funding support |  Israel, Israel,  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2022 Journal: Nat Chem Biol / Year: 2022Title: A counter-enzyme complex regulates glutamate metabolism in Bacillus subtilis. Authors: Vijay Jayaraman / D John Lee / Nadav Elad / Shay Vimer / Michal Sharon / James S Fraser / Dan S Tawfik /   Abstract: Multi-enzyme assemblies composed of metabolic enzymes catalyzing sequential reactions are being increasingly studied. Here, we report the discovery of a 1.6 megadalton multi-enzyme complex from ...Multi-enzyme assemblies composed of metabolic enzymes catalyzing sequential reactions are being increasingly studied. Here, we report the discovery of a 1.6 megadalton multi-enzyme complex from Bacillus subtilis composed of two enzymes catalyzing opposite ('counter-enzymes') rather than sequential reactions: glutamate synthase (GltAB) and glutamate dehydrogenase (GudB), which make and break glutamate, respectively. In vivo and in vitro studies show that the primary role of complex formation is to inhibit the activity of GudB. Using cryo-electron microscopy, we elucidated the structure of the complex and the molecular basis of inhibition of GudB by GltAB. The complex exhibits unusual oscillatory progress curves and is necessary for both planktonic growth, in glutamate-limiting conditions, and for biofilm growth, in glutamate-rich media. The regulation of a key metabolic enzyme by complexing with its counter enzyme may thus enable cell growth under fluctuating glutamate concentrations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23825.map.gz emd_23825.map.gz | 61.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23825-v30.xml emd-23825-v30.xml emd-23825.xml emd-23825.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23825_fsc.xml emd_23825_fsc.xml | 19.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_23825.png emd_23825.png | 190 KB | ||
| Masks |  emd_23825_msk_1.map emd_23825_msk_1.map | 67 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23825.cif.gz emd-23825.cif.gz | 7.8 KB | ||
| Others |  emd_23825_additional_1.map.gz emd_23825_additional_1.map.gz emd_23825_half_map_1.map.gz emd_23825_half_map_1.map.gz emd_23825_half_map_2.map.gz emd_23825_half_map_2.map.gz | 61.9 MB 62.3 MB 62.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23825 http://ftp.pdbj.org/pub/emdb/structures/EMD-23825 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23825 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23825 | HTTPS FTP |
-Validation report
| Summary document |  emd_23825_validation.pdf.gz emd_23825_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23825_full_validation.pdf.gz emd_23825_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_23825_validation.xml.gz emd_23825_validation.xml.gz | 19.8 KB | Display | |
| Data in CIF |  emd_23825_validation.cif.gz emd_23825_validation.cif.gz | 26.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23825 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23825 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23825 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23825 | HTTPS FTP |
-Related structure data
| Related structure data |  7mftMC  7mfmC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23825.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23825.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single GudB-GltAB asymmetric unit from GudB6-GltA6B6 complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
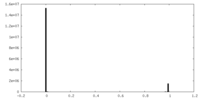
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23825_msk_1.map emd_23825_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
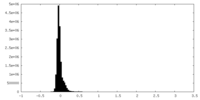
| Density Histograms |
-Additional map: Single GudB-GltAB asymmetric unit from GudB6-GltA6B6 complex. Sharpen...
| File | emd_23825_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single GudB-GltAB asymmetric unit from GudB6-GltA6B6 complex. Sharpen map | ||||||||||||
| Projections & Slices |
| ||||||||||||
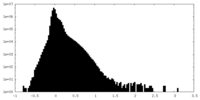
| Density Histograms |
-Half map: Single GudB-GltAB asymmetric unit from GudB6-GltA6B6 complex
| File | emd_23825_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single GudB-GltAB asymmetric unit from GudB6-GltA6B6 complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
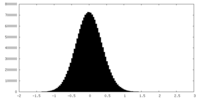
| Density Histograms |
-Half map: Single GudB-GltAB asymmetric unit from GudB6-GltA6B6 complex
| File | emd_23825_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single GudB-GltAB asymmetric unit from GudB6-GltA6B6 complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GudB-GltA-GltB
| Entire | Name: GudB-GltA-GltB |
|---|---|
| Components |
|
-Supramolecule #1: GudB-GltA-GltB
| Supramolecule | Name: GudB-GltA-GltB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Asymmetric unit (GudB-GltA-GltB) from a full GudB6-GltA6-GltB6 complex. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.62 MDa |
-Macromolecule #1: Glutamate dehydrogenase
| Macromolecule | Name: Glutamate dehydrogenase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.100715 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAADRNTGHT EEDKLDVLKS TQTVIHKALE KLGYPEEVYE LLKEPMRLLT VKIPVRMDDG SVKIFTGYRA QHNDSVGPTK GGIRFHPNV TEKEVKALSI WMSLKCGIID LPYGGGKGGI VCDPRDMSFR ELERLSRGYV RAISQIVGPT KDVPAPDVFT N SQIMAWMM ...String: MAADRNTGHT EEDKLDVLKS TQTVIHKALE KLGYPEEVYE LLKEPMRLLT VKIPVRMDDG SVKIFTGYRA QHNDSVGPTK GGIRFHPNV TEKEVKALSI WMSLKCGIID LPYGGGKGGI VCDPRDMSFR ELERLSRGYV RAISQIVGPT KDVPAPDVFT N SQIMAWMM DEYSRIDEFN SPGFITGKPL VLGGSHGRES ATAKGVTICI KEAAKKRGID IKGARVVVQG FGNAGSYLAK FM HDAGAKV VGISDAYGGL YDPEGLDIDY LLDRRDSFGT VTKLFNDTIT NQELLELDCD ILVPAAIENQ ITEENAHNIR AKI VVEAAN GPTTLEGTKI LSDRDILLVP DVLASAGGVT VSYFEWVQNN QGFYWSEEEV EEKLEKMMVK SFNNIYEMAN NRRI DMRLA AYMVGVRKMA EASRFRGWI UniProtKB: Glutamate dehydrogenase |
-Macromolecule #2: Glutamate synthase (NADPH) large chain
| Macromolecule | Name: Glutamate synthase (NADPH) large chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 169.006375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTYNQMPKAQ GLYRPEFEHD ACGIGLYAHL KGKQTHDIVK QGLKMLCQLD HRGGQGSDPD TGDGAGLLVQ IPDAFFRKEC KNINLPEKE RYGVGMVFFS QKEDERKKIE KQINALIEQE GQVVLGWRTV PVNVGKIGTV AQKSCPFVRQ VFIGASSDLK D NLSFERKL ...String: MTYNQMPKAQ GLYRPEFEHD ACGIGLYAHL KGKQTHDIVK QGLKMLCQLD HRGGQGSDPD TGDGAGLLVQ IPDAFFRKEC KNINLPEKE RYGVGMVFFS QKEDERKKIE KQINALIEQE GQVVLGWRTV PVNVGKIGTV AQKSCPFVRQ VFIGASSDLK D NLSFERKL YVIRKQAENW GVTEGLDFYF ASLSSQTIVY KGLLTPEQVD AFYSDLQDEA FVSAFALVHS RFSTNTFPTW ER AHPNRYL VHNGEINTLR GNINWMRARE QQFVSESFGE DLNKILPILN ADGSDSSILD NAFEFFVMAG RKPAHTAMML IPE PWTENT HMSKEKRAFY EYHSSLMEPW DGPTAISFTD GKQIGAILDR NGLRPARYYV TKDDYIIFSS EVGVIEVEQE NVLY KNRLE PGKMLLIDLE EGRIISDEEV KTQIATEYPY QKWLEEELVQ VNPDPESREE EQFSDLLTRQ KAFGYTYEDI QKYLI PVIK EGKDPLGSMG NDAPLAVLSD RAQSLFNYFK QLFAQVTNPP IDAIREQLVT STMTWLGAEG DLLHPSERNV RRIKLY TPV LSNEQFYALK TIVHPDLKSQ KIDVLFSEDL ERGLKDMFTQ AEKAISQGVS LLILSDKKMN ERLTPIPPLL AVSALHQ HL IRKGLRTKVS IIVESGEARE VHHFAALIGY GADAINPYLA YATYKQEIDE GRLDISYEEA VSKYGKSITE GVVKVMSK M GISTVQSYRG AQIFEAVGIS RDVIDRYFSG TASQLGGIDL QTIAEEAQRR HREAYQDDYS KTLEPGSDFQ WRNGGEHHA FNPKTIHTLQ WACRRNDYNL FKQYTKAADE ERIGFLRNLF AFDGNRKPLK LEEVESAESI VKRFKTGAMS FGSLSKEAHE ALAIAMNRL GGKSNSGEGG EDPKRFVPDE NGDDRRSAIK QIASGRFGVK SHYLVNADEL QIKMAQGAKP GEGGQLPGNK V YPWVADVR GSTPGVGLIS PPPHHDIYSI EDLAQLIHDL KNANRDARIS VKLVSKAGVG TIAAGVAKAT ADVIVISGYD GG TGASPKT SIKHTGLPWE LGLAEAHQTL MLNGLRDRVV LETDGKLMTG RDVVMAALLG AEEFGFATAP LVVLGCVMMR ACH LDTCPV GVATQNPELR KKFMGDPDHI VNYMLFIAEE VREYMAALGF KTFDEMIGRT DVLHVSERAK EHWKASQLDL STLL YQPEG VRTFQSPQNH KIDQSLDITT ILPAVQEAIE SGKEADISIE INNTNRVAGT ITGSEISKRY GEEGLPEDTI KLHFT GSAG QSFGAFVPKG MTLYLDGDSN DYVGKGLSGG KIIVKSSEGF NSASDDNVII GNVAFYGATS GEAYINGRAG ERFAVR NSG VNVVVEGIGD HGCEYMTGGS VVVLGDVGKN FAAGMSGGIA YVLTEDVKAF KRKCNLEMIL FESLEDEKEI QQIKAML ER HTAYTNSQKA EDLLDQWEDS VKKFVKVIPK NYKQMLASIE EQKAAGLSDE EAIMFAFEAN TKPKQNTAAS GQKQAVVQ UniProtKB: Glutamate synthase (NADPH) large chain |
-Macromolecule #3: Glutamate synthase (NADPH) small chain
| Macromolecule | Name: Glutamate synthase (NADPH) small chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 58.010598 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGKPTGFMEI KREKPAERDP LTRLKDWKEY SAPFSEEASK RQGARCMDCG TPFCQIGADI NGFTSGCPIY NLIPEWNGLV YRGRWKEAL ERLLKTNNFP EFTGRVCPAP CEGSCTLAIS DPAVSIKNIE RTIIDKGFEN GWIQPRIPKK RTGKKVAIVG S GPAGLASA ...String: MGKPTGFMEI KREKPAERDP LTRLKDWKEY SAPFSEEASK RQGARCMDCG TPFCQIGADI NGFTSGCPIY NLIPEWNGLV YRGRWKEAL ERLLKTNNFP EFTGRVCPAP CEGSCTLAIS DPAVSIKNIE RTIIDKGFEN GWIQPRIPKK RTGKKVAIVG S GPAGLASA DQLNQAGHSV TVFERADRAG GLLTYGIPNM KLEKGIVERR IKLLTQEGID FVTNTEIGVD ITADELKEQF DA VILCTGA QKQRDLLIEG RDSKGVHYAM DYLTLATKSY LDSNFKDKQF IDAKGKDVIV IGGGDTGADC VATALRQKAK SVH QFGKHP KLPPARTNDN MWPEQPHVFT LEYAYEEAEA KFGRDPREYS IQTTKMVADK NGKLKELHTI QMEKVKNEHG KYEF RELPG TEKVWPAQLV FIAIGFEGTE QPLLKQFGVN SVNNKISAAY GDYQTNIDGV FAAGDARRGQ SLIVWAINEG REVAR EVDR YLMGSSVLPG SWSHPQFEKG GGSGGGSGGS AWSHPQFENK UniProtKB: Glutamate synthase (NADPH) small chain |
-Macromolecule #4: FLAVIN MONONUCLEOTIDE
| Macromolecule | Name: FLAVIN MONONUCLEOTIDE / type: ligand / ID: 4 / Number of copies: 1 / Formula: FMN |
|---|---|
| Molecular weight | Theoretical: 456.344 Da |
| Chemical component information |  ChemComp-FMN: |
-Macromolecule #5: FE3-S4 CLUSTER
| Macromolecule | Name: FE3-S4 CLUSTER / type: ligand / ID: 5 / Number of copies: 1 / Formula: F3S |
|---|---|
| Molecular weight | Theoretical: 295.795 Da |
| Chemical component information |  ChemComp-F3S: |
-Macromolecule #6: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 6 / Number of copies: 1 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Macromolecule #7: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 7 / Number of copies: 2 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.9 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 47.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Details | Real-space refinement involved iterative rounds of ISOLDE, Coot and PHENIX refinement. 1OFD and 6S6T were used as templates for homology modeling in SWISS-MODEL. | ||||||||
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||||
| Output model |  PDB-7mft: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)