[English] 日本語
 Yorodumi
Yorodumi- EMDB-23788: CryoEM structure of SARS-CoV-2 RBD in complex with nanobodies Nb2... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23788 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
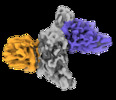
| Title | CryoEM structure of SARS-CoV-2 RBD in complex with nanobodies Nb21 and Nb105 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 Receptor binding domain nanobody / VIRAL PROTEIN / VIRAL PROTEIN-Immune System complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
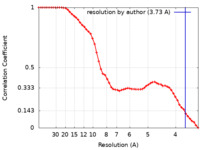
| Method | single particle reconstruction / cryo EM / Resolution: 3.73 Å | |||||||||
 Authors Authors | Huang W / Taylor DJ | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting diverse and conserved epitopes. Authors: Dapeng Sun / Zhe Sang / Yong Joon Kim / Yufei Xiang / Tomer Cohen / Anna K Belford / Alexis Huet / James F Conway / Ji Sun / Derek J Taylor / Dina Schneidman-Duhovny / Cheng Zhang / Wei Huang / Yi Shi /   Abstract: Interventions against variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are urgently needed. Stable and potent nanobodies (Nbs) that target the receptor binding domain (RBD) of ...Interventions against variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are urgently needed. Stable and potent nanobodies (Nbs) that target the receptor binding domain (RBD) of SARS-CoV-2 spike are promising therapeutics. However, it is unknown if Nbs broadly neutralize circulating variants. We found that RBD Nbs are highly resistant to variants of concern (VOCs). High-resolution cryoelectron microscopy determination of eight Nb-bound structures reveals multiple potent neutralizing epitopes clustered into three classes: Class I targets ACE2-binding sites and disrupts host receptor binding. Class II binds highly conserved epitopes and retains activity against VOCs and RBD. Cass III recognizes unique epitopes that are likely inaccessible to antibodies. Systematic comparisons of neutralizing antibodies and Nbs provided insights into how Nbs target the spike to achieve high-affinity and broadly neutralizing activity. Structure-function analysis of Nbs indicates a variety of antiviral mechanisms. Our study may guide the rational design of pan-coronavirus vaccines and therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23788.map.gz emd_23788.map.gz | 645.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23788-v30.xml emd-23788-v30.xml emd-23788.xml emd-23788.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23788_fsc.xml emd_23788_fsc.xml | 5.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_23788.png emd_23788.png | 99.4 KB | ||
| Filedesc metadata |  emd-23788.cif.gz emd-23788.cif.gz | 6 KB | ||
| Others |  emd_23788_half_map_1.map.gz emd_23788_half_map_1.map.gz emd_23788_half_map_2.map.gz emd_23788_half_map_2.map.gz | 13.9 MB 13.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23788 http://ftp.pdbj.org/pub/emdb/structures/EMD-23788 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23788 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23788 | HTTPS FTP |
-Related structure data
| Related structure data |  7me7MC  7mdwC  7mejC  7n9tC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23788.map.gz / Format: CCP4 / Size: 689.5 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23788.map.gz / Format: CCP4 / Size: 689.5 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.685 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_23788_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
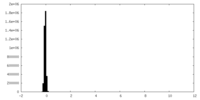
| Density Histograms |
-Half map: #1
| File | emd_23788_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
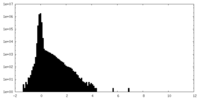
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM map of SARS-CoV-2 RBD in complex with Nb21 and Nb105
| Entire | Name: CryoEM map of SARS-CoV-2 RBD in complex with Nb21 and Nb105 |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM map of SARS-CoV-2 RBD in complex with Nb21 and Nb105
| Supramolecule | Name: CryoEM map of SARS-CoV-2 RBD in complex with Nb21 and Nb105 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.776381 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TNLCPFGEVF NATRFASVYA WNRKRISNCV ADYSVLYNSA SFSTFKCYGV SPTKLNDLCF TNVYADSFVI RGDEVRQIAP GQTGKIADY NYKLPDDFTG CVIAWNSNNL DSKVGGNYNY LYRLFRKSNL KPFERDISTE IYQAGSTPCN GVEGFNCYFP L QSYGFQPT ...String: TNLCPFGEVF NATRFASVYA WNRKRISNCV ADYSVLYNSA SFSTFKCYGV SPTKLNDLCF TNVYADSFVI RGDEVRQIAP GQTGKIADY NYKLPDDFTG CVIAWNSNNL DSKVGGNYNY LYRLFRKSNL KPFERDISTE IYQAGSTPCN GVEGFNCYFP L QSYGFQPT NGVGYQPYRV VVLSFELLHA PATVCG UniProtKB: Spike glycoprotein |
-Macromolecule #2: Nanobody Nb17
| Macromolecule | Name: Nanobody Nb17 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.54084 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HVQLVESGGG LVQAGGSLRL SCAASGSIFS SNAMSWYRQA PGKQRELVAS ITSGGNADYA DSVKGRFTIS RDKNTVYPEM SSLKPADTA VYYCHAVGQE ASAYAPRAYW GQGTQVTVSS |
-Macromolecule #3: Nanobody Nb105
| Macromolecule | Name: Nanobody Nb105 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.795319 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VQLVESGGGL VQAGGSLRLS CAVSGRTFST YGMAWFRQAP GKERDFVATI TRSGETTLYA DSVKGRFTIS RDNAKNTVYL QMNSLKIED TAVYYCAVRR DSSWGYSRDL FEYDYWGQGT QVTVS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Sugar embedding | Material: vitrified ice |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)