+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23349 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM Structure of KdpFABC in E1 state | |||||||||
 Map data Map data | Sharpened map from Non-Uniform Refinement in cryoSPARC | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationP-type K+ transporter / P-type potassium transmembrane transporter activity / potassium:proton antiporter complex / potassium ion-transporting ATPase complex / monoatomic cation transmembrane transport / potassium ion binding / potassium ion transmembrane transport / potassium ion transport / magnesium ion binding / ATP hydrolysis activity ...P-type K+ transporter / P-type potassium transmembrane transporter activity / potassium:proton antiporter complex / potassium ion-transporting ATPase complex / monoatomic cation transmembrane transport / potassium ion binding / potassium ion transmembrane transport / potassium ion transport / magnesium ion binding / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
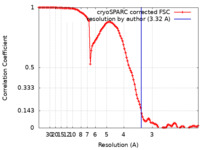
| Method | single particle reconstruction / cryo EM / Resolution: 3.32 Å | |||||||||
 Authors Authors | Sweet ME / Larsen C / Pedersen BP / Stokes DL | |||||||||
| Funding support |  United States, United States,  Denmark, 2 items Denmark, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Structural basis for potassium transport in prokaryotes by KdpFABC. Authors: Marie E Sweet / Casper Larsen / Xihui Zhang / Michael Schlame / Bjørn P Pedersen / David L Stokes /   Abstract: KdpFABC is an oligomeric K transport complex in prokaryotes that maintains ionic homeostasis under stress conditions. The complex comprises a channel-like subunit (KdpA) from the superfamily of K ...KdpFABC is an oligomeric K transport complex in prokaryotes that maintains ionic homeostasis under stress conditions. The complex comprises a channel-like subunit (KdpA) from the superfamily of K transporters and a pump-like subunit (KdpB) from the superfamily of P-type ATPases. Recent structural work has defined the architecture and generated contradictory hypotheses for the transport mechanism. Here, we use substrate analogs to stabilize four key intermediates in the reaction cycle and determine the corresponding structures by cryogenic electron microscopy. We find that KdpB undergoes conformational changes consistent with other representatives from the P-type superfamily, whereas KdpA, KdpC, and KdpF remain static. We observe a series of spherical densities that we assign as K or water and which define a pathway for K transport. This pathway runs through an intramembrane tunnel in KdpA and delivers ions to sites in the membrane domain of KdpB. Our structures suggest a mechanism where ATP hydrolysis is coupled to K transfer between alternative sites in KdpB, ultimately reaching a low-affinity site where a water-filled pathway allows release of K to the cytoplasm. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23349.map.gz emd_23349.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23349-v30.xml emd-23349-v30.xml emd-23349.xml emd-23349.xml | 24.9 KB 24.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23349_fsc.xml emd_23349_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_23349.png emd_23349.png | 88.5 KB | ||
| Others |  emd_23349_half_map_1.map.gz emd_23349_half_map_1.map.gz emd_23349_half_map_2.map.gz emd_23349_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23349 http://ftp.pdbj.org/pub/emdb/structures/EMD-23349 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23349 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23349 | HTTPS FTP |
-Validation report
| Summary document |  emd_23349_validation.pdf.gz emd_23349_validation.pdf.gz | 579.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23349_full_validation.pdf.gz emd_23349_full_validation.pdf.gz | 579.4 KB | Display | |
| Data in XML |  emd_23349_validation.xml.gz emd_23349_validation.xml.gz | 18.9 KB | Display | |
| Data in CIF |  emd_23349_validation.cif.gz emd_23349_validation.cif.gz | 24.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23349 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23349 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23349 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23349 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23349.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23349.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map from Non-Uniform Refinement in cryoSPARC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
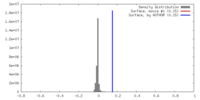
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Un-sharpened half-map A from Non-Uniform Refinement in cryoSPARC
| File | emd_23349_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Un-sharpened half-map A from Non-Uniform Refinement in cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
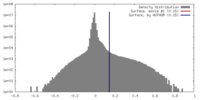
| Density Histograms |
-Half map: Un-sharpened half-map B from Non-Uniform Refinement in cryoSPARC
| File | emd_23349_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Un-sharpened half-map B from Non-Uniform Refinement in cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : KdpFABC in E1 state with VO4 and K+ in sample buffer
| Entire | Name: KdpFABC in E1 state with VO4 and K+ in sample buffer |
|---|---|
| Components |
|
-Supramolecule #1: KdpFABC in E1 state with VO4 and K+ in sample buffer
| Supramolecule | Name: KdpFABC in E1 state with VO4 and K+ in sample buffer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: KdpFABC complex solublized in decyl maltopyranoside (DM) detergent. |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 160 KDa |
-Macromolecule #1: Potassium-transporting ATPase potassium-binding subunit
| Macromolecule | Name: Potassium-transporting ATPase potassium-binding subunit type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MAAQGFLLIA TFLLVLMVLA RPLGSGLARL INDIPLPGTT GVERVLFRAL GVSDREMNWK QYLCAILGLN MLGLAVLFFM LLGQHYLPL NPQQLPGLSW DLALNTAVSF VTNTNWRSYS GETTLSYFSQ MAGLTVQNFL SAASGIAVIF ALIRAFTRQS M STLGNAWV ...String: MAAQGFLLIA TFLLVLMVLA RPLGSGLARL INDIPLPGTT GVERVLFRAL GVSDREMNWK QYLCAILGLN MLGLAVLFFM LLGQHYLPL NPQQLPGLSW DLALNTAVSF VTNTNWRSYS GETTLSYFSQ MAGLTVQNFL SAASGIAVIF ALIRAFTRQS M STLGNAWV DLLRITLWVL VPVALLIALF FIQQGALQNF LPYQAVNTVE GAQQLLPMGP VASQEAIKML GTNGGGFFNA NS SHPFENP TALTNFVQML AIFLIPTALC FAFGEVMGDR RQGRMLLWAM SVIFVICVGV VMWAEVQGNP HLLALGTDSS INM EGKESR FGVLVSSLFA VVTTAASCGA VIAMHDSFTA LGGMVPMWLM QIGEVVFGGV GSGLYGMMLF VLLAVFIAGL MIGR TPEYL GKKIDVREMK LTALAILVTP TLVLMGAALA MMTDAGRSAM LNPGPHGFSE VLYAVSSAAN NNGSAFAGLS ANSPF WNCL LAFCMFVGRF GVIIPVMAIA GSLVSKKSQA ASSGTLPTHG PLFVGLLIGT VLLVGALTFI PALALGPVAE YLS |
-Macromolecule #2: Potassium-transporting ATPase ATP-binding subunit
| Macromolecule | Name: Potassium-transporting ATPase ATP-binding subunit / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO / EC number: P-type K+ transporter |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MSRKQLALFE PTLVVQALKE AVKKLNPQAQ WRNPVMFIVW IGSLLTTCIS IAMASGAMPG NALFSAAISG WLWITVLFAN FAEALAEGR SKAQANSLKG VKKTAFARKL REPKYGAAAD KVPADQLRKG DIVLVEAGDI IPCDGEVIEG GASVDESAIT G EAAPVIRE ...String: MSRKQLALFE PTLVVQALKE AVKKLNPQAQ WRNPVMFIVW IGSLLTTCIS IAMASGAMPG NALFSAAISG WLWITVLFAN FAEALAEGR SKAQANSLKG VKKTAFARKL REPKYGAAAD KVPADQLRKG DIVLVEAGDI IPCDGEVIEG GASVDESAIT G EAAPVIRE SGGDFASVTG GTRILSDWLV IECSVNPGET FLDRMIAMVE GAQRRKTPNE IALTILLIAL TIVFLLATAT LW PFSAWGG NAVSVTVLVA LLVCLIPTTI GGLLSAIGVA GMSRMLGANV IATSGRAVEA AGDVDVLLLD KTGTITLGNR QAS EFIPAQ GVDEKTLADA AQLASLADET PEGRSIVILA KQRFNLRERD VQSLHATFVP FTAQSRMSGI NIDNRMIRKG SVDA IRRHV EANGGHFPTD VDQKVDQVAR QGATPLVVVE GSRVLGVIAL KDIVKGGIKE RFAQLRKMGI KTVMITGDNR LTAAA IAAE AGVDDFLAEA TPEAKLALIR QYQAEGRLVA MTGDGTNDAP ALAQADVAVA MNSGTQAAKE AGNMVDLDSN PTKLIE VVH IGKQMLMTRG SLTTFSIAND VAKYFAIIPA AFAATYPQLN ALNIMCLHSP DSAILSAVIF NALIIVFLIP LALKGVS YK PLTASAMLRR NLWIYGLGGL LVPFIGIKVI DLLLTVCGLV |
-Macromolecule #3: Potassium-transporting ATPase KdpC subunit
| Macromolecule | Name: Potassium-transporting ATPase KdpC subunit / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MSGLRPALST FIFLLLITGG VYPLLTTVLG QWWFPWQANG SLIREGDTVR GSALIGQNFT GNGYFHGRPS ATAEMPYNPQ ASGGSNLAV SNPELDKLIA ARVAALRAAN PDASASVPVE LVTASASGLD NNITPQAAAW QIPRVAKARN LSVEQLTQLI A KYSQQPLV ...String: MSGLRPALST FIFLLLITGG VYPLLTTVLG QWWFPWQANG SLIREGDTVR GSALIGQNFT GNGYFHGRPS ATAEMPYNPQ ASGGSNLAV SNPELDKLIA ARVAALRAAN PDASASVPVE LVTASASGLD NNITPQAAAW QIPRVAKARN LSVEQLTQLI A KYSQQPLV KYIGQPVVNI VELNLALDKL DEGTGLVPRG SSHHHHHHHH |
-Macromolecule #4: Potassium-transporting ATPase KdpF subunit
| Macromolecule | Name: Potassium-transporting ATPase KdpF subunit / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MSAGVITGVL LVFLLLGYLV YALINAEAF |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.5 mg/mL | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: VO4 was added to the protein after size-exclusion chromatography and incubated 1 hr prior to freezing | ||||||||||||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 50.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR Details: PELCO easiGlow Glow Discharge Cleaning System used for glow discharge with default settings. | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 3 uL of protein mixture applied to grid. Blot time 4 seconds, blot force 0, no wait before plunging.. | ||||||||||||||||||||||||
| Details | DM-solublized KdpFABC complex purified through SEC the same day as grid freezing. Peak fraction diluted to 4.5 mg/mL and complexed with VO4 for 1 hr at room temperature prior to application to grid. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 77.0 K / Max: 77.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Digitization - Sampling interval: 5.0 µm / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 3265 / Average exposure time: 2.7 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Correlation coefficient |
|---|
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)