+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22237 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPitc-3nt | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transcriptional factor / TRANSCRIPTION / promoter / multidrug recognition | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Yang Y / Liu C | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural visualization of transcription activated by a multidrug-sensing MerR family regulator. Authors: Yang Yang / Chang Liu / Wei Zhou / Wei Shi / Ming Chen / Baoyue Zhang / David G Schatz / Yangbo Hu / Bin Liu /   Abstract: Bacterial RNA polymerase (RNAP) holoenzyme initiates transcription by recognizing the conserved -35 and -10 promoter elements that are optimally separated by a 17-bp spacer. The MerR family of ...Bacterial RNA polymerase (RNAP) holoenzyme initiates transcription by recognizing the conserved -35 and -10 promoter elements that are optimally separated by a 17-bp spacer. The MerR family of transcriptional regulators activate suboptimal 19-20 bp spacer promoters in response to myriad cellular signals, ranging from heavy metals to drug-like compounds. The regulation of transcription by MerR family regulators is not fully understood. Here we report one crystal structure of a multidrug-sensing MerR family regulator EcmrR and nine cryo-electron microscopy structures that capture the EcmrR-dependent transcription process from promoter opening to initial transcription to RNA elongation. These structures reveal that EcmrR is a dual ligand-binding factor that reshapes the suboptimal 19-bp spacer DNA to enable optimal promoter recognition, sustains promoter remodeling to stabilize initial transcribing complexes, and finally dissociates from the promoter to reverse DNA remodeling and facilitate the transition to elongation. Our findings yield a comprehensive model for transcription regulation by MerR family factors and provide insights into the transition from transcription initiation to elongation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22237.map.gz emd_22237.map.gz | 197.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22237-v30.xml emd-22237-v30.xml emd-22237.xml emd-22237.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22237.png emd_22237.png | 318.2 KB | ||
| Filedesc metadata |  emd-22237.cif.gz emd-22237.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22237 http://ftp.pdbj.org/pub/emdb/structures/EMD-22237 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22237 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22237 | HTTPS FTP |
-Validation report
| Summary document |  emd_22237_validation.pdf.gz emd_22237_validation.pdf.gz | 558.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22237_full_validation.pdf.gz emd_22237_full_validation.pdf.gz | 558.2 KB | Display | |
| Data in XML |  emd_22237_validation.xml.gz emd_22237_validation.xml.gz | 6.9 KB | Display | |
| Data in CIF |  emd_22237_validation.cif.gz emd_22237_validation.cif.gz | 8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22237 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22237 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22237 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22237 | HTTPS FTP |
-Related structure data
| Related structure data |  6xlaMC  6wl5C  6xl5C  6xl6C  6xl9C  6xljC  6xlkC  6xllC  6xlmC  6xlnC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22237.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22237.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.895 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
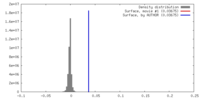
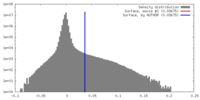
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : EcmrR-spacer DNA complex in EcmrR-RNAP-promoter initial transcrib...
| Entire | Name: EcmrR-spacer DNA complex in EcmrR-RNAP-promoter initial transcribing complex with 3-nt RNA transcript (EcmrR-RPitc-3nt) |
|---|---|
| Components |
|
-Supramolecule #1: EcmrR-spacer DNA complex in EcmrR-RNAP-promoter initial transcrib...
| Supramolecule | Name: EcmrR-spacer DNA complex in EcmrR-RNAP-promoter initial transcribing complex with 3-nt RNA transcript (EcmrR-RPitc-3nt) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: MerR family transcriptional regulator EcmrR
| Macromolecule | Name: MerR family transcriptional regulator EcmrR / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.394191 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SQIGLFSKIC RVTIKTLHYY NKIGLLVPAY INPDNGYRFY TSDQLMKFHQ IASLRQLGFT ITEIVTLTQD ENSCHIIERR RLEIQKQIR DMADMLSRIN HYLQHKKKER IMLYQAALKE IPECIVYSKR FIVPDFSSYI KLIPPIGQEV MKANPGLTLT T PAYCFTLY ...String: SQIGLFSKIC RVTIKTLHYY NKIGLLVPAY INPDNGYRFY TSDQLMKFHQ IASLRQLGFT ITEIVTLTQD ENSCHIIERR RLEIQKQIR DMADMLSRIN HYLQHKKKER IMLYQAALKE IPECIVYSKR FIVPDFSSYI KLIPPIGQEV MKANPGLTLT T PAYCFTLY HDKEYKEKNM DVEFCEAVND FGKNEGNIIF QVIPAITAVT VIHKGPYDSL RNAYIYLMQW VEDNGYLLTN SP RESYIDG IWNKQDSAEW MTEIQFPVEK V |
-Macromolecule #2: synthetic non-template strand DNA (54-MER)
| Macromolecule | Name: synthetic non-template strand DNA (54-MER) / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.718658 KDa |
| Sequence | String: (DG)(DC)(DC)(DT)(DT)(DG)(DA)(DC)(DC)(DC) (DT)(DC)(DC)(DC)(DC)(DT)(DA)(DA)(DG)(DG) (DG)(DG)(DA)(DG)(DG)(DG)(DT)(DT)(DT) (DA)(DG)(DA)(DT)(DT)(DG)(DT)(DG)(DT)(DG) (DC) (DA)(DG)(DT)(DC)(DT)(DG) ...String: (DG)(DC)(DC)(DT)(DT)(DG)(DA)(DC)(DC)(DC) (DT)(DC)(DC)(DC)(DC)(DT)(DA)(DA)(DG)(DG) (DG)(DG)(DA)(DG)(DG)(DG)(DT)(DT)(DT) (DA)(DG)(DA)(DT)(DT)(DG)(DT)(DG)(DT)(DG) (DC) (DA)(DG)(DT)(DC)(DT)(DG)(DA)(DC) (DG)(DC)(DG)(DG)(DC)(DG) |
-Macromolecule #3: synthetic template strand DNA (54-MER)
| Macromolecule | Name: synthetic template strand DNA (54-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.634643 KDa |
| Sequence | String: (DC)(DG)(DC)(DC)(DG)(DC)(DG)(DT)(DC)(DA) (DG)(DA)(DC)(DT)(DC)(DG)(DT)(DA)(DG)(DG) (DA)(DA)(DT)(DC)(DT)(DA)(DA)(DA)(DC) (DC)(DC)(DT)(DC)(DC)(DC)(DC)(DT)(DT)(DA) (DG) (DG)(DG)(DG)(DA)(DG)(DG) ...String: (DC)(DG)(DC)(DC)(DG)(DC)(DG)(DT)(DC)(DA) (DG)(DA)(DC)(DT)(DC)(DG)(DT)(DA)(DG)(DG) (DA)(DA)(DT)(DC)(DT)(DA)(DA)(DA)(DC) (DC)(DC)(DT)(DC)(DC)(DC)(DC)(DT)(DT)(DA) (DG) (DG)(DG)(DG)(DA)(DG)(DG)(DG)(DT) (DC)(DA)(DA)(DG)(DG)(DC) |
-Macromolecule #4: TETRAPHENYLANTIMONIUM ION
| Macromolecule | Name: TETRAPHENYLANTIMONIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: 118 |
|---|---|
| Molecular weight | Theoretical: 430.176 Da |
| Chemical component information | 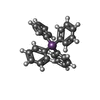 ChemComp-118: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Average exposure time: 30.0 sec. / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)