+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22189 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | R. capsulatus cyt bc1 (CIII2) at 3.3A | ||||||||||||||||||
 Map data Map data | CIII2 | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | cytochrome bc1 membrane protein complex ubiquinone:cytochrome c oxidoreductase Complex III / OXIDOREDUCTASE | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationrespiratory chain complex III / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / respiratory electron transport chain / 2 iron, 2 sulfur cluster binding / electron transfer activity / heme binding / metal ion binding / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Rhodobacter capsulatus SB 1003 (bacteria) / Rhodobacter capsulatus SB 1003 (bacteria) /  Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (bacteria) Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (bacteria) | ||||||||||||||||||
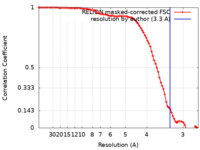
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||||||||
 Authors Authors | Steimle S / Van Eeuwen T | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Cryo-EM structures of engineered active bc-cbb type CIIICIV super-complexes and electronic communication between the complexes. Authors: Stefan Steimle / Trevor van Eeuwen / Yavuz Ozturk / Hee Jong Kim / Merav Braitbard / Nur Selamoglu / Benjamin A Garcia / Dina Schneidman-Duhovny / Kenji Murakami / Fevzi Daldal /    Abstract: Respiratory electron transport complexes are organized as individual entities or combined as large supercomplexes (SC). Gram-negative bacteria deploy a mitochondrial-like cytochrome (cyt) bc (Complex ...Respiratory electron transport complexes are organized as individual entities or combined as large supercomplexes (SC). Gram-negative bacteria deploy a mitochondrial-like cytochrome (cyt) bc (Complex III, CIII), and may have specific cbb-type cyt c oxidases (Complex IV, CIV) instead of the canonical aa-type CIV. Electron transfer between these complexes is mediated by soluble (c) and membrane-anchored (c) cyts. Here, we report the structure of an engineered bc-cbb type SC (CIIICIV, 5.2 Å resolution) and three conformers of native CIII (3.3 Å resolution). The SC is active in vivo and in vitro, contains all catalytic subunits and cofactors, and two extra transmembrane helices attributed to cyt c and the assembly factor CcoH. The cyt c is integral to SC, its cyt domain is mobile and it conveys electrons to CIV differently than cyt c. The successful production of a native-like functional SC and determination of its structure illustrate the characteristics of membrane-confined and membrane-external respiratory electron transport pathways in Gram-negative bacteria. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22189.map.gz emd_22189.map.gz | 4.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22189-v30.xml emd-22189-v30.xml emd-22189.xml emd-22189.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22189_fsc.xml emd_22189_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_22189.png emd_22189.png | 244.3 KB | ||
| Filedesc metadata |  emd-22189.cif.gz emd-22189.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22189 http://ftp.pdbj.org/pub/emdb/structures/EMD-22189 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22189 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22189 | HTTPS FTP |
-Related structure data
| Related structure data |  6xi0MC  6xktC  6xkuC  6xkvC  6xkwC  6xkxC  6xkzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10735 (Title: Cryo-EM structures of engineered active bc1-cbb3 type CIII2CIV super-complexes and electronic communication between the complexes EMPIAR-10735 (Title: Cryo-EM structures of engineered active bc1-cbb3 type CIII2CIV super-complexes and electronic communication between the complexesData size: 35.8 TB Data #1: tripartite SC - dataset 1 [micrographs - multiframe] Data #2: tripartite SC - dataset 2 [micrographs - multiframe] Data #3: tripartite SC - dataset 3 [micrographs - multiframe] Data #4: tripartite SC - dataset 4 [micrographs - multiframe] Data #5: tripartite SC - dataset 5 [micrographs - multiframe] Data #6: tripartite SC - dataset 6 [micrographs - multiframe] Data #7: tripartite SC - dataset 7 [micrographs - multiframe] Data #8: bipartite SC - dataset 1 [micrographs - single frame] Data #9: bipartite SC - dataset 2 [micrographs - multiframe] Data #10: bipartite SC - dataset 3 [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22189.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22189.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CIII2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : homodimer of ubiquinol:cytochrome c reductase (complex III or cyt...
| Entire | Name: homodimer of ubiquinol:cytochrome c reductase (complex III or cyt bc1 complex) |
|---|---|
| Components |
|
-Supramolecule #1: homodimer of ubiquinol:cytochrome c reductase (complex III or cyt...
| Supramolecule | Name: homodimer of ubiquinol:cytochrome c reductase (complex III or cyt bc1 complex) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Rhodobacter capsulatus SB 1003 (bacteria) Rhodobacter capsulatus SB 1003 (bacteria) |
| Molecular weight | Theoretical: 250 KDa |
-Macromolecule #1: Ubiquinol-cytochrome c reductase iron-sulfur subunit
| Macromolecule | Name: Ubiquinol-cytochrome c reductase iron-sulfur subunit / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: quinol-cytochrome-c reductase |
|---|---|
| Source (natural) | Organism:  Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (bacteria) Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (bacteria)Strain: ATCC BAA-309 / NBRC 16581 / SB1003 |
| Molecular weight | Theoretical: 20.465109 KDa |
| Recombinant expression | Organism:  Rhodobacter capsulatus SB 1003 (bacteria) Rhodobacter capsulatus SB 1003 (bacteria) |
| Sequence | String: MSHAEDNAGT RRDFLYHATA ATGVVVTGAA VWPLINQMNA SADVKAMASI FVDVSAVEVG TQLTVKWRGK PVFIRRRDEK DIELARSVP LGALRDTSAE NANKPGAEAT DENRTLPAFD GTNTGEWLVM LGVCTHLGCV PMGDKSGDFG GWFCPCHGSH Y DSAGRIRK GPAPRNLDIP VAAFVDETTI KLG UniProtKB: Ubiquinol-cytochrome c reductase iron-sulfur subunit |
-Macromolecule #2: Cytochrome b
| Macromolecule | Name: Cytochrome b / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (bacteria) Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (bacteria)Strain: ATCC BAA-309 / NBRC 16581 / SB1003 |
| Molecular weight | Theoretical: 49.386469 KDa |
| Recombinant expression | Organism:  Rhodobacter capsulatus SB 1003 (bacteria) Rhodobacter capsulatus SB 1003 (bacteria) |
| Sequence | String: MSGIPHDHYE PKTGIEKWLH DRLPIVGLVY DTIMIPTPKN LNWWWIWGIV LAFTLVLQIV TGIVLAMHYT PHVDLAFASV EHIMRDVNG GWAMRYIHAN GASLFFLAVY IHIFRGLYYG SYKAPREITW IVGMVIYLLM MGTAFMGYVL PWGQMSFWGA T VITGLFGA ...String: MSGIPHDHYE PKTGIEKWLH DRLPIVGLVY DTIMIPTPKN LNWWWIWGIV LAFTLVLQIV TGIVLAMHYT PHVDLAFASV EHIMRDVNG GWAMRYIHAN GASLFFLAVY IHIFRGLYYG SYKAPREITW IVGMVIYLLM MGTAFMGYVL PWGQMSFWGA T VITGLFGA IPGIGPSIQA WLLGGPAVDN ATLNRFFSLH YLLPFVIAAL VAIHIWAFHT TGNNNPTGVE VRRTSKADAE KD TLPFWPY FVIKDLFALA LVLLGFFAVV AYMPNYLGHP DNYVQANPLS TPAHIVPEWY FLPFYAILRA FAADVWVVIL VDG LTFGIV DAKFFGVIAM FGAIAVMALA PWLDTSKVRS GAYRPKFRMW FWFLVLDFVV LTWVGAMPTE YPYDWISLIA STYW FAYFL VILPLLGATE KPEPIPASIE EDFNSHYGNP AE UniProtKB: Cytochrome b |
-Macromolecule #3: Cytochrome c1
| Macromolecule | Name: Cytochrome c1 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (bacteria) Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (bacteria)Strain: ATCC BAA-309 / NBRC 16581 / SB1003 |
| Molecular weight | Theoretical: 30.352615 KDa |
| Recombinant expression | Organism:  Rhodobacter capsulatus SB 1003 (bacteria) Rhodobacter capsulatus SB 1003 (bacteria) |
| Sequence | String: MKKLLISAVS ALVLGSGAAF ANSNVPDHAF SFEGIFGKYD QAQLRRGFQV YNEVCSACHG MKFVPIRTLA DDGGPQLDPT FVREYAAGL DTIIDKDSGE ERDRKETDMF PTRVGDGMGP DLSVMAKARA GFSGPAGSGM NQLFKGMGGP EYIYNYVIGF E ENPECAPE ...String: MKKLLISAVS ALVLGSGAAF ANSNVPDHAF SFEGIFGKYD QAQLRRGFQV YNEVCSACHG MKFVPIRTLA DDGGPQLDPT FVREYAAGL DTIIDKDSGE ERDRKETDMF PTRVGDGMGP DLSVMAKARA GFSGPAGSGM NQLFKGMGGP EYIYNYVIGF E ENPECAPE GIDGYYYNKT FQIGGVPDTC KDAAGVKITH GSWARMPPPL VDDQVTYEDG TPATVDQMAQ DVSAFLMWAA EP KLVARKQ MGLVAMVMLG LLSVMLYLTN KRLWAPYKGH KA UniProtKB: Cytochrome c1 |
-Macromolecule #4: FE2/S2 (INORGANIC) CLUSTER
| Macromolecule | Name: FE2/S2 (INORGANIC) CLUSTER / type: ligand / ID: 4 / Number of copies: 2 / Formula: FES |
|---|---|
| Molecular weight | Theoretical: 175.82 Da |
| Chemical component information |  ChemComp-FES: |
-Macromolecule #5: HEME C
| Macromolecule | Name: HEME C / type: ligand / ID: 5 / Number of copies: 6 / Formula: HEC |
|---|---|
| Molecular weight | Theoretical: 618.503 Da |
| Chemical component information |  ChemComp-HEC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)