+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22009 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human TRPA1 in complex with agonist GNE551 | |||||||||
 Map data Map data | Density-modified map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRPA1 / channel / agonist / MEMBRANE PROTEIN / MEMBRANE PROTEIN-Agonist complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of blood circulation / positive regulation of monoatomic anion transport / cellular response to food / temperature-gated cation channel activity / regulation of neuronal action potential / cellular response to carbon dioxide / osmolarity-sensing monoatomic cation channel activity / stereocilium bundle / detection of chemical stimulus involved in sensory perception of pain / urinary bladder smooth muscle contraction ...regulation of blood circulation / positive regulation of monoatomic anion transport / cellular response to food / temperature-gated cation channel activity / regulation of neuronal action potential / cellular response to carbon dioxide / osmolarity-sensing monoatomic cation channel activity / stereocilium bundle / detection of chemical stimulus involved in sensory perception of pain / urinary bladder smooth muscle contraction / thermoception / cellular response to toxic substance / cellular response to caffeine / response to pain / TRP channels / calcium ion transmembrane import into cytosol / cellular response to cold / intracellularly gated calcium channel activity / detection of mechanical stimulus involved in sensory perception of pain / positive regulation of insulin secretion involved in cellular response to glucose stimulus / voltage-gated calcium channel activity / monoatomic ion transport / sensory perception of pain / response to cold / calcium ion transmembrane transport / calcium channel activity / cellular response to hydrogen peroxide / intracellular calcium ion homeostasis / cellular response to heat / channel activity / protein homotetramerization / cell surface receptor signaling pathway / apical plasma membrane / response to xenobiotic stimulus / axon / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
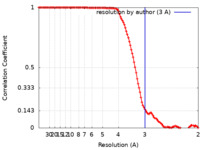
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Rohou A / Rouge L / Chen H | |||||||||
 Citation Citation |  Journal: Neuron / Year: 2021 Journal: Neuron / Year: 2021Title: A Non-covalent Ligand Reveals Biased Agonism of the TRPA1 Ion Channel. Authors: Chang Liu / Rebecca Reese / Simon Vu / Lionel Rougé / Shannon D Shields / Satoko Kakiuchi-Kiyota / Huifen Chen / Kevin Johnson / Yu Patrick Shi / Tania Chernov-Rogan / Demi Maria Zabala ...Authors: Chang Liu / Rebecca Reese / Simon Vu / Lionel Rougé / Shannon D Shields / Satoko Kakiuchi-Kiyota / Huifen Chen / Kevin Johnson / Yu Patrick Shi / Tania Chernov-Rogan / Demi Maria Zabala Greiner / Pawan Bir Kohli / David Hackos / Bobby Brillantes / Christine Tam / Tianbo Li / Jianyong Wang / Brian Safina / Steve Magnuson / Matthew Volgraf / Jian Payandeh / Jie Zheng / Alexis Rohou / Jun Chen /  Abstract: The TRPA1 ion channel is activated by electrophilic compounds through the covalent modification of intracellular cysteine residues. How non-covalent agonists activate the channel and whether covalent ...The TRPA1 ion channel is activated by electrophilic compounds through the covalent modification of intracellular cysteine residues. How non-covalent agonists activate the channel and whether covalent and non-covalent agonists elicit the same physiological responses are not understood. Here, we report the discovery of a non-covalent agonist, GNE551, and determine a cryo-EM structure of the TRPA1-GNE551 complex, revealing a distinct binding pocket and ligand-interaction mechanism. Unlike the covalent agonist allyl isothiocyanate, which elicits channel desensitization, tachyphylaxis, and transient pain, GNE551 activates TRPA1 into a distinct conducting state without desensitization and induces persistent pain. Furthermore, GNE551-evoked pain is relatively insensitive to antagonist treatment. Thus, we demonstrate the biased agonism of TRPA1, a finding that has important implications for the discovery of effective drugs tailored to different disease etiologies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22009.map.gz emd_22009.map.gz | 5.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22009-v30.xml emd-22009-v30.xml emd-22009.xml emd-22009.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22009_fsc.xml emd_22009_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_22009.png emd_22009.png | 155.3 KB | ||
| Filedesc metadata |  emd-22009.cif.gz emd-22009.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22009 http://ftp.pdbj.org/pub/emdb/structures/EMD-22009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22009 | HTTPS FTP |
-Related structure data
| Related structure data |  6x2jMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22009.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22009.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density-modified map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TRPA1 bound by agonist GNE551
| Entire | Name: TRPA1 bound by agonist GNE551 |
|---|---|
| Components |
|
-Supramolecule #1: TRPA1 bound by agonist GNE551
| Supramolecule | Name: TRPA1 bound by agonist GNE551 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transient receptor potential cation channel subfamily A member 1
| Macromolecule | Name: Transient receptor potential cation channel subfamily A member 1 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.622125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SPLHFAASYG RINTCQRLLQ DISDTRLLNE GDLHGMTPLH LAAKNGHDKV VQLLLKKGAL FLSDHNGWTA LHHASMGGYT QTMKVILDT NLKCTDRLDE DGNTALHFAA REGHAKAVAL LLSHNADIVL NKQQASFLHL ALHNKRKEVV LTIIRSKRWD E CLKIFSHN ...String: SPLHFAASYG RINTCQRLLQ DISDTRLLNE GDLHGMTPLH LAAKNGHDKV VQLLLKKGAL FLSDHNGWTA LHHASMGGYT QTMKVILDT NLKCTDRLDE DGNTALHFAA REGHAKAVAL LLSHNADIVL NKQQASFLHL ALHNKRKEVV LTIIRSKRWD E CLKIFSHN SPGNKCPITE MIEYLPECMK VLLDFCMLHS TEDKSCRDYY IEYNFKYLQC PLEFTKKTPT QDVIYEPLTA LN AMVQNNR IELLNHPVCK EYLLMKWLAY GFRAHMMNLG SYCLGLIPMT ILVVNIKPGM AFNSTGIINE TSDHSEILDT TNS YLIKTC MILVFLSSIF GYCKEAGQIF QQKRNYFMDI SNVLEWIIYT TGIIFVLPLF VEIPAHLQWQ CGAIAVYFYW MNFL LYLQR FENCGIFIVM LEVILKTLLR STVVFIFLLL AFGLSFYILL NLQDPFSSPL LSIIQTFSMM LGDINYRESF LEPYL RNEL AHPVLSFAQL VSFTIFVPIV LMNLLIGLAV GDIADVQKHA SLKRIAMQVE LHTSLEKKLP LWFLRKVDQK STIVYP NKP RSGGMLFHIF CFLFCTGEIR QEIPNADKSL EMEILKQKYR LKDLTFLLEK QHELIKLIIQ KMEIISET UniProtKB: Transient receptor potential cation channel subfamily A member 1 |
-Macromolecule #2: 5-amino-1-[(4-bromo-2-fluorophenyl)methyl]-N-(2,5-dimethoxyphenyl...
| Macromolecule | Name: 5-amino-1-[(4-bromo-2-fluorophenyl)methyl]-N-(2,5-dimethoxyphenyl)-1H-1,2,3-triazole-4-carboxamide type: ligand / ID: 2 / Number of copies: 4 / Formula: ULJ |
|---|---|
| Molecular weight | Theoretical: 450.262 Da |
| Chemical component information |  ChemComp-ULJ: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.33 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.2 Component:
| ||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 300 / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: HOLEY / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: GOLD / Support film - #1 - topology: HOLEY / Support film - #1 - Film thickness: 25 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 0.1 kPa / Details: Solarus plasma cleaner | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Triple blot. Put blot in vitroblot Apply 3.5ul to grid, wait 30sec, blot manually Apply 3.5ul to grid, wait 30sec, blot manually, apply final 3.5ul, final blot by vitrobot (3.5s). |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 11063 / Average exposure time: 1.6 sec. / Average electron dose: 41.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)