[English] 日本語
 Yorodumi
Yorodumi- EMDB-21635: Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA pro... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21635 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure VI-B) | ||||||||||||
 Map data Map data | Map VI-B blocfilt filtered and with B-factor -50 applied | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Ribosome / EF-Tu / tRNA | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationstringent response / ornithine decarboxylase inhibitor activity / transcription antitermination factor activity, RNA binding / misfolded RNA binding / Group I intron splicing / RNA folding / transcriptional attenuation / endoribonuclease inhibitor activity / positive regulation of ribosome biogenesis / RNA-binding transcription regulator activity ...stringent response / ornithine decarboxylase inhibitor activity / transcription antitermination factor activity, RNA binding / misfolded RNA binding / Group I intron splicing / RNA folding / transcriptional attenuation / endoribonuclease inhibitor activity / positive regulation of ribosome biogenesis / RNA-binding transcription regulator activity / translational termination / negative regulation of cytoplasmic translation / four-way junction DNA binding / DnaA-L2 complex / translation repressor activity / negative regulation of translational initiation / regulation of mRNA stability / negative regulation of DNA-templated DNA replication initiation / mRNA regulatory element binding translation repressor activity / positive regulation of RNA splicing / assembly of large subunit precursor of preribosome / cytosolic ribosome assembly / response to reactive oxygen species / regulation of DNA-templated transcription elongation / ribosome assembly / transcription elongation factor complex / transcription antitermination / DNA endonuclease activity / regulation of cell growth / DNA-templated transcription termination / response to radiation / maintenance of translational fidelity / mRNA 5'-UTR binding / regulation of translation / large ribosomal subunit / ribosome biogenesis / transferase activity / ribosome binding / ribosomal small subunit biogenesis / ribosomal small subunit assembly / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / response to antibiotic / negative regulation of DNA-templated transcription / mRNA binding / DNA binding / RNA binding / zinc ion binding / membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |    | ||||||||||||
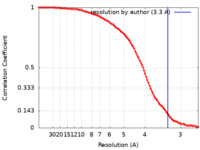
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||
 Authors Authors | Loveland AB / Demo G | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading. Authors: Anna B Loveland / Gabriel Demo / Andrei A Korostelev /   Abstract: Ribosomes accurately decode mRNA by proofreading each aminoacyl-tRNA that is delivered by the elongation factor EF-Tu. To understand the molecular mechanism of this proofreading step it is necessary ...Ribosomes accurately decode mRNA by proofreading each aminoacyl-tRNA that is delivered by the elongation factor EF-Tu. To understand the molecular mechanism of this proofreading step it is necessary to visualize GTP-catalysed elongation, which has remained a challenge. Here we use time-resolved cryogenic electron microscopy to reveal 33 ribosomal states after the delivery of aminoacyl-tRNA by EF-Tu•GTP. Instead of locking cognate tRNA upon initial recognition, the ribosomal decoding centre dynamically monitors codon-anticodon interactions before and after GTP hydrolysis. GTP hydrolysis enables the GTPase domain of EF-Tu to extend away, releasing EF-Tu from tRNA. The 30S subunit then locks cognate tRNA in the decoding centre and rotates, enabling the tRNA to bypass 50S protrusions during accommodation into the peptidyl transferase centre. By contrast, the decoding centre fails to lock near-cognate tRNA, enabling the dissociation of near-cognate tRNA both during initial selection (before GTP hydrolysis) and proofreading (after GTP hydrolysis). These findings reveal structural similarity between ribosomes in initial selection states and in proofreading states, which together govern the efficient rejection of incorrect tRNA. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21635.map.gz emd_21635.map.gz | 85.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21635-v30.xml emd-21635-v30.xml emd-21635.xml emd-21635.xml | 77.7 KB 77.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21635_fsc.xml emd_21635_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_21635.png emd_21635.png | 143.8 KB | ||
| Filedesc metadata |  emd-21635.cif.gz emd-21635.cif.gz | 13.5 KB | ||
| Others |  emd_21635_additional.map.gz emd_21635_additional.map.gz emd_21635_half_map_1.map.gz emd_21635_half_map_1.map.gz emd_21635_half_map_2.map.gz emd_21635_half_map_2.map.gz | 84.5 MB 34 MB 34 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21635 http://ftp.pdbj.org/pub/emdb/structures/EMD-21635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21635 | HTTPS FTP |
-Validation report
| Summary document |  emd_21635_validation.pdf.gz emd_21635_validation.pdf.gz | 891.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21635_full_validation.pdf.gz emd_21635_full_validation.pdf.gz | 891.3 KB | Display | |
| Data in XML |  emd_21635_validation.xml.gz emd_21635_validation.xml.gz | 18.1 KB | Display | |
| Data in CIF |  emd_21635_validation.cif.gz emd_21635_validation.cif.gz | 23.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21635 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21635 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21635 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21635 | HTTPS FTP |
-Related structure data
| Related structure data |  6wdgMC  6wd0C  6wd1C  6wd2C  6wd3C  6wd4C  6wd5C  6wd6C  6wd7C  6wd8C  6wd9C  6wdaC  6wdbC  6wdcC  6wddC  6wdeC  6wdfC  6wdhC  6wdiC  6wdjC  6wdkC  6wdlC  6wdmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21635.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21635.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map VI-B blocfilt filtered and with B-factor -50 applied | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.333 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Map VI-B
| File | emd_21635_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map VI-B | ||||||||||||
| Projections & Slices |
| ||||||||||||
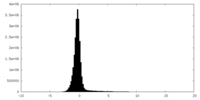
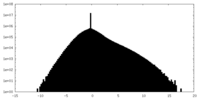
| Density Histograms |
-Half map: Half map 2 VI-B
| File | emd_21635_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 VI-B | ||||||||||||
| Projections & Slices |
| ||||||||||||
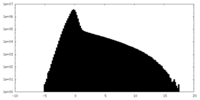
| Density Histograms |
-Half map: Half map 1 VI-B
| File | emd_21635_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 VI-B | ||||||||||||
| Projections & Slices |
| ||||||||||||
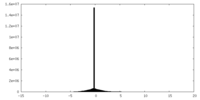
| Density Histograms |
- Sample components
Sample components
+Entire : Map VI-B
+Supramolecule #1: Map VI-B
+Macromolecule #1: 50S ribosomal protein L2
+Macromolecule #2: 50S ribosomal protein L3
+Macromolecule #3: 50S ribosomal protein L4
+Macromolecule #4: 50S ribosomal protein L5
+Macromolecule #5: 50S ribosomal protein L6
+Macromolecule #6: 50S ribosomal protein L9
+Macromolecule #7: 50S ribosomal protein L10
+Macromolecule #8: 50S ribosomal protein L11
+Macromolecule #9: 50S ribosomal protein L13
+Macromolecule #10: 50S ribosomal protein L14
+Macromolecule #11: 50S ribosomal protein L15
+Macromolecule #12: 50S ribosomal protein L16
+Macromolecule #13: 50S ribosomal protein L17
+Macromolecule #14: 50S ribosomal protein L18
+Macromolecule #15: 50S ribosomal protein L19
+Macromolecule #16: 50S ribosomal protein L20
+Macromolecule #17: 50S ribosomal protein L21
+Macromolecule #18: 50S ribosomal protein L22
+Macromolecule #19: 50S ribosomal protein L23
+Macromolecule #20: 50S ribosomal protein L24
+Macromolecule #21: 50S ribosomal protein L25
+Macromolecule #22: 50S ribosomal protein L27
+Macromolecule #23: 50S ribosomal protein L28
+Macromolecule #24: 50S ribosomal protein L29
+Macromolecule #25: 50S ribosomal protein L30
+Macromolecule #26: 50S ribosomal protein L32
+Macromolecule #27: 50S ribosomal protein L33
+Macromolecule #28: 50S ribosomal protein L34
+Macromolecule #29: 50S ribosomal protein L35
+Macromolecule #30: 50S ribosomal protein L36
+Macromolecule #31: 30S ribosomal protein S2
+Macromolecule #32: 30S ribosomal protein S3
+Macromolecule #33: 30S ribosomal protein S4
+Macromolecule #34: 30S ribosomal protein S5
+Macromolecule #35: 30S ribosomal protein S6
+Macromolecule #36: 30S ribosomal protein S7
+Macromolecule #37: 30S ribosomal protein S8
+Macromolecule #38: 30S ribosomal protein S9
+Macromolecule #39: 30S ribosomal protein S10
+Macromolecule #40: 30S ribosomal protein S11
+Macromolecule #41: 30S ribosomal protein S12
+Macromolecule #42: 30S ribosomal protein S13
+Macromolecule #43: 30S ribosomal protein S14
+Macromolecule #44: 30S ribosomal protein S15
+Macromolecule #45: 30S ribosomal protein S16
+Macromolecule #46: 30S ribosomal protein S17
+Macromolecule #47: 30S ribosomal protein S18
+Macromolecule #48: 30S ribosomal protein S19
+Macromolecule #49: 30S ribosomal protein S20
+Macromolecule #50: 30S ribosomal protein S21
+Macromolecule #51: 50S ribosomal protein L1
+Macromolecule #52: 16S ribosomal RNA
+Macromolecule #53: 23S ribosomal RNA
+Macromolecule #54: 5S ribosomal RNA
+Macromolecule #55: tRNAfMet
+Macromolecule #56: mRNA
+Macromolecule #57: tRNAPhe
+Macromolecule #58: PHENYLALANINE
+Macromolecule #59: N-FORMYLMETHIONINE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-35 / Number grids imaged: 1 / Number real images: 3218 / Average exposure time: 1.0 sec. / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-6wdg: |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)