+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2156 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of axonemal dynein-c in the ADP.Vi state | |||||||||
 Map data Map data | Reconstruction of axonemal dynein-c prepared in the ADP.Vi state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Dynein / motor protein / microtubule / cytoskeleton / cilia / flagella / axoneme / AAA+ ATPase / cryo-electron microscopy | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 22.0 Å | |||||||||
 Authors Authors | Roberts AJ / Malkova B / Walker ML / Sakakibara H / Numata N / Kon T / Ohkura R / Edwards TA / Knight PJ / Sutoh K ...Roberts AJ / Malkova B / Walker ML / Sakakibara H / Numata N / Kon T / Ohkura R / Edwards TA / Knight PJ / Sutoh K / Oiwa K / Burgess SA | |||||||||
 Citation Citation |  Journal: Structure / Year: 2012 Journal: Structure / Year: 2012Title: ATP-driven remodeling of the linker domain in the dynein motor. Authors: Anthony J Roberts / Bara Malkova / Matt L Walker / Hitoshi Sakakibara / Naoki Numata / Takahide Kon / Reiko Ohkura / Thomas A Edwards / Peter J Knight / Kazuo Sutoh / Kazuhiro Oiwa / Stan A Burgess /  Abstract: Dynein ATPases are the largest known cytoskeletal motors and perform critical functions in cells: carrying cargo along microtubules in the cytoplasm and powering flagellar beating. Dyneins are ...Dynein ATPases are the largest known cytoskeletal motors and perform critical functions in cells: carrying cargo along microtubules in the cytoplasm and powering flagellar beating. Dyneins are members of the AAA+ superfamily of ring-shaped enzymes, but how they harness this architecture to produce movement is poorly understood. Here, we have used cryo-EM to determine 3D maps of native flagellar dynein-c and a cytoplasmic dynein motor domain in different nucleotide states. The structures show key sites of conformational change within the AAA+ ring and a large rearrangement of the "linker" domain, involving a hinge near its middle. Analysis of a mutant in which the linker "undocks" from the ring indicates that linker remodeling requires energy that is supplied by interactions with the AAA+ modules. Fitting the dynein-c structures into flagellar tomograms suggests how this mechanism could drive sliding between microtubules, and also has implications for cytoplasmic cargo transport. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2156.map.gz emd_2156.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2156-v30.xml emd-2156-v30.xml emd-2156.xml emd-2156.xml | 8.2 KB 8.2 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2156.png EMD-2156.png emd_2156.png emd_2156.png | 60.1 KB 60.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2156 http://ftp.pdbj.org/pub/emdb/structures/EMD-2156 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2156 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2156 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2156.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2156.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of axonemal dynein-c prepared in the ADP.Vi state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.16 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
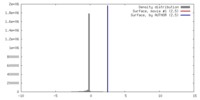
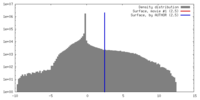
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Axonemal dynein-c prepared in the ADP.Vi state
| Entire | Name: Axonemal dynein-c prepared in the ADP.Vi state |
|---|---|
| Components |
|
-Supramolecule #1000: Axonemal dynein-c prepared in the ADP.Vi state
| Supramolecule | Name: Axonemal dynein-c prepared in the ADP.Vi state / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Macromolecule #1: Dynein-c
| Macromolecule | Name: Dynein-c / type: protein_or_peptide / ID: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.19 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 10 mM MOPS, 3 mM MgCl2, 1 mM EGTA |
| Grid | Details: Quantifoil or lacey carbon film, glow discharged |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | May 1, 2006 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 228 / Average electron dose: 25 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 69444 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Phase flipping |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 22.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Spider / Number images used: 22667 |
| Final two d classification | Number classes: 799 |
 Movie
Movie Controller
Controller






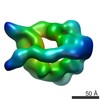

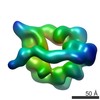



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)