+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20912 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Group 1 H1 influenza stem nanoparticle with an H38R mutation | |||||||||
 Map data Map data | Map after postprocessing | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Influenza A virus Influenza A virus | |||||||||
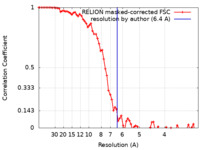
| Method | single particle reconstruction / cryo EM / Resolution: 6.4 Å | |||||||||
 Authors Authors | Tsybovsky Y / Stephens T / Kanekiyo M / Graham BS | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Glycan repositioning of influenza hemagglutinin stem facilitates the elicitation of protective cross-group antibody responses. Authors: Seyhan Boyoglu-Barnum / Geoffrey B Hutchinson / Jeffrey C Boyington / Syed M Moin / Rebecca A Gillespie / Yaroslav Tsybovsky / Tyler Stephens / John R Vaile / Julia Lederhofer / Kizzmekia S ...Authors: Seyhan Boyoglu-Barnum / Geoffrey B Hutchinson / Jeffrey C Boyington / Syed M Moin / Rebecca A Gillespie / Yaroslav Tsybovsky / Tyler Stephens / John R Vaile / Julia Lederhofer / Kizzmekia S Corbett / Brian E Fisher / Hadi M Yassine / Sarah F Andrews / Michelle C Crank / Adrian B McDermott / John R Mascola / Barney S Graham / Masaru Kanekiyo /  Abstract: The conserved hemagglutinin (HA) stem has been a focus of universal influenza vaccine efforts. Influenza A group 1 HA stem-nanoparticles have been demonstrated to confer heterosubtypic protection in ...The conserved hemagglutinin (HA) stem has been a focus of universal influenza vaccine efforts. Influenza A group 1 HA stem-nanoparticles have been demonstrated to confer heterosubtypic protection in animals; however, the protection does not extend to group 2 viruses, due in part to differences in glycosylation between group 1 and 2 stems. Here, we show that introducing the group 2 glycan at Asn38 to a group 1 stem-nanoparticle (gN38 variant) based on A/New Caledonia/20/99 (H1N1) broadens antibody responses to cross-react with group 2 HAs. Immunoglobulins elicited by the gN38 variant provide complete protection against group 2 H7N9 virus infection, while the variant loses protection against a group 1 H5N1 virus. The N38 glycan thus is pivotal in directing antibody responses by controlling access to group-determining stem epitopes. Precise targeting of stem-directed antibody responses to the site of vulnerability by glycan repositioning may be a step towards achieving cross-group influenza protection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20912.map.gz emd_20912.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20912-v30.xml emd-20912-v30.xml emd-20912.xml emd-20912.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20912_fsc.xml emd_20912_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_20912.png emd_20912.png | 103.6 KB | ||
| Masks |  emd_20912_msk_1.map emd_20912_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Others |  emd_20912_additional.map.gz emd_20912_additional.map.gz emd_20912_additional_1.map.gz emd_20912_additional_1.map.gz emd_20912_half_map_1.map.gz emd_20912_half_map_1.map.gz emd_20912_half_map_2.map.gz emd_20912_half_map_2.map.gz | 20.3 MB 20.3 MB 20.7 MB 20.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20912 http://ftp.pdbj.org/pub/emdb/structures/EMD-20912 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20912 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20912 | HTTPS FTP |
-Validation report
| Summary document |  emd_20912_validation.pdf.gz emd_20912_validation.pdf.gz | 78.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20912_full_validation.pdf.gz emd_20912_full_validation.pdf.gz | 78 KB | Display | |
| Data in XML |  emd_20912_validation.xml.gz emd_20912_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20912 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20912 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20912 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20912 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20912.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20912.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map after postprocessing | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.58 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20912_msk_1.map emd_20912_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
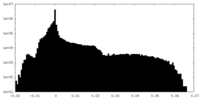
| Density Histograms |
-Additional map: Map before postprocessing
| File | emd_20912_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map before postprocessing | ||||||||||||
| Projections & Slices |
| ||||||||||||
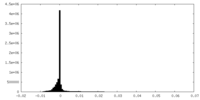
| Density Histograms |
-Additional map: Map before postprocessing
| File | emd_20912_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map before postprocessing | ||||||||||||
| Projections & Slices |
| ||||||||||||
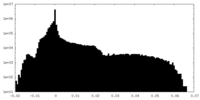
| Density Histograms |
-Half map: Half-map 1
| File | emd_20912_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
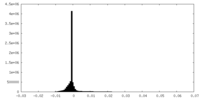
| Density Histograms |
-Half map: Half-map 2
| File | emd_20912_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : H1ssF R38
| Entire | Name: H1ssF R38 |
|---|---|
| Components |
|
-Supramolecule #1: H1ssF R38
| Supramolecule | Name: H1ssF R38 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Ferritin-based nanoparticle displaying Influenza hemagglutinin stems |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: 293 Expi Homo sapiens (human) / Recombinant cell: 293 Expi |
| Molecular weight | Theoretical: 1.031 MDa |
-Macromolecule #1: H1ssF R38
| Macromolecule | Name: H1ssF R38 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKAKLLVLLC TFTATYADTI CIGYHANNST DTVDTVLEKN VTVTRSVNLG SGLRMVTGLR NIPQRETRGL FGAIAGFIEG GWTGMVDGWY GYHHQNEQGS GYAADQKSTQ NAINGITNMV NSVIEKMGSG GSGTDLAELL VLLLNERTLD FHDSNVKNLY EKVKSQLKNN ...String: MKAKLLVLLC TFTATYADTI CIGYHANNST DTVDTVLEKN VTVTRSVNLG SGLRMVTGLR NIPQRETRGL FGAIAGFIEG GWTGMVDGWY GYHHQNEQGS GYAADQKSTQ NAINGITNMV NSVIEKMGSG GSGTDLAELL VLLLNERTLD FHDSNVKNLY EKVKSQLKNN AKEIGNGCFE FYHKCNNECM ESVKNGTYDY PKYSEESKLN REKIDSGGDI IKLLNEQVNK EMQSSNLYMS MSSWCYTHSL DGAGLFLFDH AAEEYEHAKK LIIFLNENNV PVQLTSISAP EHKFEGLTQI FQKAYEHEQH ISESINNIVD HAIKSKDHAT FNFLQWYVAE QHEEEVLFKD ILDKIELIGN ENHGLYLADQ YVKGIAKSRK SGS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: PBS |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 297 K / Instrument: FEI VITROBOT MARK IV / Details: Drop volume: 2.7 microliters. |
- Electron microscopy
Electron microscopy
| Microscope | TFS TALOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 106 / Average exposure time: 2.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 92000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)