+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20537 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
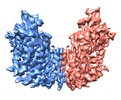
| Title | cryo-EM structure of human NKCC1 | |||||||||
 Map data Map data | Final map, sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NKCC1 / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of cell volume / positive regulation of aspartate secretion / transepithelial ammonium transport / regulation of matrix metallopeptidase secretion / cell body membrane / metal ion transmembrane transporter activity / inorganic anion import across plasma membrane / inorganic cation import across plasma membrane / chloride:monoatomic cation symporter activity / sodium:potassium:chloride symporter activity ...positive regulation of cell volume / positive regulation of aspartate secretion / transepithelial ammonium transport / regulation of matrix metallopeptidase secretion / cell body membrane / metal ion transmembrane transporter activity / inorganic anion import across plasma membrane / inorganic cation import across plasma membrane / chloride:monoatomic cation symporter activity / sodium:potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / potassium ion transmembrane transporter activity / transepithelial chloride transport / intracellular chloride ion homeostasis / ammonium transmembrane transport / negative regulation of vascular wound healing / sodium ion homeostasis / ammonium channel activity / chloride ion homeostasis / cell projection membrane / cellular response to chemokine / T cell chemotaxis / potassium ion homeostasis / intracellular sodium ion homeostasis / sodium ion import across plasma membrane / cell volume homeostasis / cellular response to potassium ion / hyperosmotic response / regulation of spontaneous synaptic transmission / gamma-aminobutyric acid signaling pathway / maintenance of blood-brain barrier / potassium ion import across plasma membrane / intracellular potassium ion homeostasis / lateral plasma membrane / transport across blood-brain barrier / monoatomic ion transport / chloride transmembrane transport / basal plasma membrane / sodium ion transmembrane transport / cytoplasmic vesicle membrane / cell periphery / cell projection / Hsp90 protein binding / extracellular vesicle / protein-folding chaperone binding / cell body / basolateral plasma membrane / neuron projection / apical plasma membrane / neuronal cell body / protein kinase binding / extracellular exosome / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.46 Å | |||||||||
 Authors Authors | Cao E / Wang Q | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structure of the human cation-chloride cotransporter NKCC1 determined by single-particle electron cryo-microscopy. Authors: Xiaoyong Yang / Qinzhe Wang / Erhu Cao /  Abstract: The secondary active cation-chloride cotransporters (CCCs) utilize the existing Na and/or K gradients to move Cl into or out of cells. NKCC1 is an intensively studied member of the CCC family and ...The secondary active cation-chloride cotransporters (CCCs) utilize the existing Na and/or K gradients to move Cl into or out of cells. NKCC1 is an intensively studied member of the CCC family and plays fundamental roles in regulating trans-epithelial ion movement, cell volume, chloride homeostasis and neuronal excitability. Here, we report a cryo-EM structure of human NKCC1 captured in a partially loaded, inward-open state. NKCC1 assembles into a dimer, with the first ten transmembrane (TM) helices harboring the transport core and TM11-TM12 helices lining the dimer interface. TM1 and TM6 helices break α-helical geometry halfway across the lipid bilayer where ion binding sites are organized around these discontinuous regions. NKCC1 may harbor multiple extracellular entryways and intracellular exits, raising the possibility that K, Na, and Cl ions may traverse along their own routes for translocation. NKCC1 structure provides a blueprint for further probing structure-function relationships of NKCC1 and other CCCs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20537.map.gz emd_20537.map.gz | 97.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20537-v30.xml emd-20537-v30.xml emd-20537.xml emd-20537.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20537.png emd_20537.png | 197 KB | ||
| Filedesc metadata |  emd-20537.cif.gz emd-20537.cif.gz | 5.7 KB | ||
| Others |  emd_20537_additional.map.gz emd_20537_additional.map.gz emd_20537_half_map_1.map.gz emd_20537_half_map_1.map.gz emd_20537_half_map_2.map.gz emd_20537_half_map_2.map.gz | 95.3 MB 95.4 MB 95.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20537 http://ftp.pdbj.org/pub/emdb/structures/EMD-20537 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20537 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20537 | HTTPS FTP |
-Related structure data
| Related structure data |  6pztMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20537.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20537.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final map, sharpened | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Final map, unsharpened
| File | emd_20537_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final map, unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
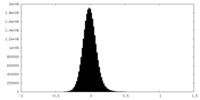
| Density Histograms |
-Half map: half map from cryosparc v2
| File | emd_20537_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map from cryosparc v2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
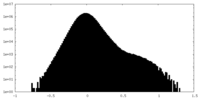
| Density Histograms |
-Half map: Another half map from cryosparc v2
| File | emd_20537_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Another half map from cryosparc v2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
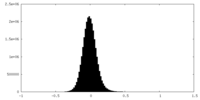
| Density Histograms |
- Sample components
Sample components
-Entire : human NKCC1
| Entire | Name: human NKCC1 |
|---|---|
| Components |
|
-Supramolecule #1: human NKCC1
| Supramolecule | Name: human NKCC1 / type: cell / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 12 member 2
| Macromolecule | Name: Solute carrier family 12 member 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 131.668484 KDa |
| Recombinant expression | Organism: Mammalian expression vector BsrGI-MCS-pcDNA3.1 (others) |
| Sequence | String: MEPRPTAPSS GAPGLAGVGE TPSAAALAAA RVELPGTAVP SVPEDAAPAS RDGGGVRDEG PAAAGDGLGR PLGPTPSQSR FQVDLVSEN AGRAAAAAAA AAAAAAAAGA GAGAKQTPAD GEASGESEPA KGSEEAKGRF RVNFVDPAAS SSAEDSLSDA A GVGVDGPN ...String: MEPRPTAPSS GAPGLAGVGE TPSAAALAAA RVELPGTAVP SVPEDAAPAS RDGGGVRDEG PAAAGDGLGR PLGPTPSQSR FQVDLVSEN AGRAAAAAAA AAAAAAAAGA GAGAKQTPAD GEASGESEPA KGSEEAKGRF RVNFVDPAAS SSAEDSLSDA A GVGVDGPN VSFQNGGDTV LSEGSSLHSG GGGGSGHHQH YYYDTHTNTY YLRTFGHNTM DAVPRIDHYR HTAAQLGEKL LR PSLAELH DELEKEPFED GFANGEESTP TRDAVVTYTA ESKGVVKFGW INGVLVRCML NIWGVMLFIR LSWIVGQAGI GLS VLVIMM ATVVTTITGL STSAIATNGF VRGGRAYYLI SRSLGPEFGG AIGLIFAFAN AVAVAMYVVG FAETVVELLK EHSI LMIDE INDIRIIGAI TVVILLGISV AGMEWEAKAQ IVLLVILLLA IGDFVIGTFI PLESKKPKGF FGYKSEIFNE NFGPD FREE ETFFSVFAIF FPAATGILAG ANISGDLADP QSAIPKGTLL AILITTLVYV GIAVSVGSCV VRDATGNVND TIVTEL TNC TSAACKLNFD FSSCESSPCS YGLMNNFQVM SMVSGFTPLI SAGIFSATLS SALASLVSAP KIFQALCKDN IYPAFQM FA KGYGKNNEPL RGYILTFLIA LGFILIAELN VIAPIISNFF LASYALINFS VFHASLAKSP GWRPAFKYYN MWISLLGA I LCCIVMFVIN WWAALLTYVI VLGLYIYVTY KKPDVNWGSS TQALTYLNAL QHSIRLSGVE DHVKNFRPQC LVMTGAPNS RPALLHLVHD FTKNVGLMIC GHVHMGPRRQ AMKEMSIDQA KYQRWLIKNK MKAFYAPVHA DDLREGAQYL MQAAGLGRMK PNTLVLGFK KDWLQADMRD VDMYINLFHD AFDIQYGVVV IRLKEGLDIS HLQGQEELLS SQEKSPGTKD VVVSVEYSKK S DLDTSKPL SEKPITHKVE EEDGKTATQP LLKKESKGPI VPLNVADQKL LEASTQFQKK QGKNTIDVWW LFDDGGLTLL IP YLLTTKK KWKDCKIRVF IGGKINRIDH DRRAMATLLS KFRIDFSDIM VLGDINTKPK KENIIAFEEI IEPYRLHEDD KEQ DIADKM KEDEPWRITD NELELYKTKT YRQIRLNELL KEHSSTANII VMSLPVARKG AVSSALYMAW LEALSKDLPP ILLV RGNHQ SVLTFYS UniProtKB: Solute carrier family 12 member 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 10.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.46 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 90803 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6pzt: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)