[English] 日本語
 Yorodumi
Yorodumi- EMDB-20241: Mobile loops and electrostatic interactions maintain the flexible... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20241 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mobile loops and electrostatic interactions maintain the flexible lambda tail tube | |||||||||
 Map data Map data | lambda tail tube | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tail tube / siphoviridae / helical / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationvirus tail, tube / symbiont genome ejection through host cell envelope, long flexible tail mechanism / viral tail assembly / host cell cytoplasm Similarity search - Function | |||||||||
| Biological species |  Escherichia phage lambda (virus) Escherichia phage lambda (virus) | |||||||||
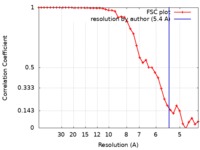
| Method | helical reconstruction / cryo EM / Resolution: 5.4 Å | |||||||||
 Authors Authors | Campbell P / Duda RL | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2020 Journal: J Mol Biol / Year: 2020Title: Mobile Loops and Electrostatic Interactions Maintain the Flexible Tail Tube of Bacteriophage Lambda. Authors: Patricia L Campbell / Robert L Duda / Jamie Nassur / James F Conway / Alexis Huet /  Abstract: The long flexible tail tube of bacteriophage lambda connects its capsid to the tail tip. On infection, a DNA ejection signal is passed from the tip, along the tube to the capsid that triggers passage ...The long flexible tail tube of bacteriophage lambda connects its capsid to the tail tip. On infection, a DNA ejection signal is passed from the tip, along the tube to the capsid that triggers passage of the DNA down the tube and into the host bacterium. The tail tube is built from repeating units of the major tail protein, gpV, which has two distinctive domains. Its N-terminal domain has the same fold as proteins that form the rigid inner tubes of contractile tail phages, such as T4, and its C-terminal domain adopt an Ig-like fold of unknown function. We determined structures of the lambda tail tube in free tails and in virions before and after DNA ejection using cryoelectron microscopy. Modeling of the density maps reveals how electrostatic interactions and a mobile loop participate in assembly and also impart flexibility to the tube while maintaining its integrity. We also demonstrate how a common protein fold produces rigid tubes in some phages but flexible tubes in others. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20241.map.gz emd_20241.map.gz | 27.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20241-v30.xml emd-20241-v30.xml emd-20241.xml emd-20241.xml | 13.3 KB 13.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20241_fsc.xml emd_20241_fsc.xml | 3.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_20241.png emd_20241.png | 133 KB | ||
| Filedesc metadata |  emd-20241.cif.gz emd-20241.cif.gz | 5.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20241 http://ftp.pdbj.org/pub/emdb/structures/EMD-20241 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20241 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20241 | HTTPS FTP |
-Validation report
| Summary document |  emd_20241_validation.pdf.gz emd_20241_validation.pdf.gz | 530 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20241_full_validation.pdf.gz emd_20241_full_validation.pdf.gz | 529.6 KB | Display | |
| Data in XML |  emd_20241_validation.xml.gz emd_20241_validation.xml.gz | 8 KB | Display | |
| Data in CIF |  emd_20241_validation.cif.gz emd_20241_validation.cif.gz | 9.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20241 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20241 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20241 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20241 | HTTPS FTP |
-Related structure data
| Related structure data |  6p3eMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20241.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20241.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | lambda tail tube | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : tail tube of the lambda phage
| Entire | Name: tail tube of the lambda phage |
|---|---|
| Components |
|
-Supramolecule #1: tail tube of the lambda phage
| Supramolecule | Name: tail tube of the lambda phage / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: plasmid expression of the tail genes |
|---|---|
| Source (natural) | Organism:  Escherichia phage lambda (virus) Escherichia phage lambda (virus) |
| Molecular weight | Theoretical: 37 kDa/nm |
-Macromolecule #1: Tail tube protein
| Macromolecule | Name: Tail tube protein / type: protein_or_peptide / ID: 1 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage lambda (virus) Escherichia phage lambda (virus) |
| Molecular weight | Theoretical: 25.831779 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPVPNPTMPV KGAGTTLWVY KGSGDPYANP LSDVDWSRLA KVKDLTPGEL TAESYDDSYL DDEDADWTAT GQGQKSAGDT SFTLAWMPG EQGQQALLAW FNEGDTRAYK IRFPNGTVDV FRGWVSSIGK AVTAKEVITR TVKVTNVGRP SMAEDRSTVT A ATGMTVTP ...String: MPVPNPTMPV KGAGTTLWVY KGSGDPYANP LSDVDWSRLA KVKDLTPGEL TAESYDDSYL DDEDADWTAT GQGQKSAGDT SFTLAWMPG EQGQQALLAW FNEGDTRAYK IRFPNGTVDV FRGWVSSIGK AVTAKEVITR TVKVTNVGRP SMAEDRSTVT A ATGMTVTP ASTSVVKGQS TTLTVAFQPE GVTDKSFRAV SADKTKATVS VSGMTITVNG VAAGKVNIPV VSGNGEFAAV AE ITVTAS UniProtKB: Tail tube protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. | ||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK II | ||||||
| Details | purified tail |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 1240 / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 200 | ||||||
| Output model |  PDB-6p3e: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)