[English] 日本語
 Yorodumi
Yorodumi- EMDB-20230: Cryo-EM structure of Escherichia coli RNAP polymerase holoenzyme -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20230 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Escherichia coli RNAP polymerase holoenzyme | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsigma factor antagonist complex / RNA polymerase complex / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / sigma factor activity / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility ...sigma factor antagonist complex / RNA polymerase complex / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / sigma factor activity / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility / nitrate assimilation / DNA-directed RNA polymerase complex / regulation of DNA-templated transcription elongation / transcription elongation factor complex / transcription antitermination / cell motility / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / response to heat / protein-containing complex assembly / intracellular iron ion homeostasis / protein dimerization activity / response to antibiotic / negative regulation of DNA-templated transcription / DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / metal ion binding / membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.05 Å | |||||||||
 Authors Authors | Chen J / Chiu CE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: TraR allosterically regulates transcription initiation by altering RNA polymerase conformation. Authors: James Chen / Saumya Gopalkrishnan / Courtney Chiu / Albert Y Chen / Elizabeth A Campbell / Richard L Gourse / Wilma Ross / Seth A Darst /  Abstract: TraR and its homolog DksA are bacterial proteins that regulate transcription initiation by binding directly to RNA polymerase (RNAP) rather than to promoter DNA. Effects of TraR mimic the combined ...TraR and its homolog DksA are bacterial proteins that regulate transcription initiation by binding directly to RNA polymerase (RNAP) rather than to promoter DNA. Effects of TraR mimic the combined effects of DksA and its cofactor ppGpp, but the structural basis for regulation by these factors remains unclear. Here, we use cryo-electron microscopy to determine structures of RNAP, with or without TraR, and of an RNAP-promoter complex. TraR binding induced RNAP conformational changes not seen in previous crystallographic analyses, and a quantitative analysis revealed TraR-induced changes in RNAP conformational heterogeneity. These changes involve mobile regions of RNAP affecting promoter DNA interactions, including the βlobe, the clamp, the bridge helix, and several lineage-specific insertions. Using mutational approaches, we show that these structural changes, as well as effects on σ region 1.1, are critical for transcription activation or inhibition, depending on the kinetic features of regulated promoters. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20230.map.gz emd_20230.map.gz | 28.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20230-v30.xml emd-20230-v30.xml emd-20230.xml emd-20230.xml | 9 KB 9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20230_fsc.xml emd_20230_fsc.xml | 8.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_20230.png emd_20230.png | 62.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20230 http://ftp.pdbj.org/pub/emdb/structures/EMD-20230 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20230 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20230 | HTTPS FTP |
-Validation report
| Summary document |  emd_20230_validation.pdf.gz emd_20230_validation.pdf.gz | 384.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20230_full_validation.pdf.gz emd_20230_full_validation.pdf.gz | 384.4 KB | Display | |
| Data in XML |  emd_20230_validation.xml.gz emd_20230_validation.xml.gz | 8.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20230 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20230 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20230 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20230 | HTTPS FTP |
-Related structure data
| Related structure data |  6p1kMC  0348C  0349C  6n57C  6n58C  6oulC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20230.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20230.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
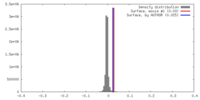
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Escherichia coli sigma70-holoenzyme
| Entire | Name: Escherichia coli sigma70-holoenzyme |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia coli sigma70-holoenzyme
| Supramolecule | Name: Escherichia coli sigma70-holoenzyme / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 0.7 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)