+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacteriophage JBD30 capsid | |||||||||
 Map data Map data | main_map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacteriophage JBD30 / virion / capsid / major capsid protein / VIRUS | |||||||||
| Function / homology | Bacteriophage Mu, GpT / Mu-like prophage major head subunit gpT / Bacteriophage Mu GpT domain-containing protein Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas phage JBD30 (virus) Pseudomonas phage JBD30 (virus) | |||||||||
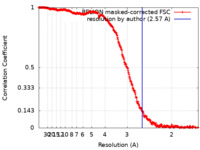
| Method | single particle reconstruction / cryo EM / Resolution: 2.57 Å | |||||||||
 Authors Authors | Valentova L / Fuzik T / Plevka P | |||||||||
| Funding support |  Czech Republic, European Union, 2 items Czech Republic, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2024 Journal: EMBO J / Year: 2024Title: Structure and replication of Pseudomonas aeruginosa phage JBD30. Authors: Lucie Valentová / Tibor Füzik / Jiří Nováček / Zuzana Hlavenková / Jakub Pospíšil / Pavel Plevka /  Abstract: Bacteriophages are the most abundant biological entities on Earth, but our understanding of many aspects of their lifecycles is still incomplete. Here, we have structurally analysed the infection ...Bacteriophages are the most abundant biological entities on Earth, but our understanding of many aspects of their lifecycles is still incomplete. Here, we have structurally analysed the infection cycle of the siphophage Casadabanvirus JBD30. Using its baseplate, JBD30 attaches to Pseudomonas aeruginosa via the bacterial type IV pilus, whose subsequent retraction brings the phage to the bacterial cell surface. Cryo-electron microscopy structures of the baseplate-pilus complex show that the tripod of baseplate receptor-binding proteins attaches to the outer bacterial membrane. The tripod and baseplate then open to release three copies of the tape-measure protein, an event that is followed by DNA ejection. JBD30 major capsid proteins assemble into procapsids, which expand by 7% in diameter upon filling with phage dsDNA. The DNA-filled heads are finally joined with 180-nm-long tails, which bend easily because flexible loops mediate contacts between the successive discs of major tail proteins. It is likely that the structural features and replication mechanisms described here are conserved among siphophages that utilize the type IV pili for initial cell attachment. #1:  Journal: Embo J. / Year: 2024 Journal: Embo J. / Year: 2024Title: Structure and replication of Pseudomonas aeruginosa phage JBD30 Authors: Valentova L / Plevka P / Fuzik T / Novacek J / Pospisil J | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19280.map.gz emd_19280.map.gz | 2.1 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19280-v30.xml emd-19280-v30.xml emd-19280.xml emd-19280.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19280_fsc.xml emd_19280_fsc.xml | 29.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_19280.png emd_19280.png | 263.6 KB | ||
| Masks |  emd_19280_msk_1.map emd_19280_msk_1.map | 2.2 GB |  Mask map Mask map | |
| Filedesc metadata |  emd-19280.cif.gz emd-19280.cif.gz | 6.1 KB | ||
| Others |  emd_19280_half_map_1.map.gz emd_19280_half_map_1.map.gz emd_19280_half_map_2.map.gz emd_19280_half_map_2.map.gz | 1.8 GB 1.8 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19280 http://ftp.pdbj.org/pub/emdb/structures/EMD-19280 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19280 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19280 | HTTPS FTP |
-Related structure data
| Related structure data |  8rknMC  8rk3C  8rk4C  8rk5C  8rk6C  8rk7C  8rk8C  8rk9C  8rkaC  8rkbC  8rkcC  8rkoC  8rkxC  8rqeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19280.map.gz / Format: CCP4 / Size: 2.2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19280.map.gz / Format: CCP4 / Size: 2.2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main_map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8336 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19280_msk_1.map emd_19280_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
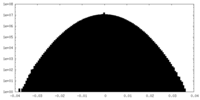
| Projections & Slices |
| ||||||||||||
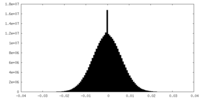
| Density Histograms |
-Half map: half map 2
| File | emd_19280_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_map_2 | ||||||||||||
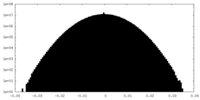
| Projections & Slices |
| ||||||||||||
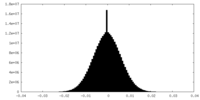
| Density Histograms |
-Half map: half map 1
| File | emd_19280_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_map_1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pseudomonas phage JBD30
| Entire | Name:  Pseudomonas phage JBD30 (virus) Pseudomonas phage JBD30 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Pseudomonas phage JBD30
| Supramolecule | Name: Pseudomonas phage JBD30 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Phage JBD30 was propagated in P. aeruginosa strain BAA-28 and purified using CsCl gradient. NCBI-ID: 1223260 / Sci species name: Pseudomonas phage JBD30 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Molecular weight | Theoretical: 14.14 MDa |
| Virus shell | Shell ID: 1 / Name: JBD30 capsid / Diameter: 640.0 Å / T number (triangulation number): 7 |
-Macromolecule #1: Bacteriophage Mu GpT domain-containing protein
| Macromolecule | Name: Bacteriophage Mu GpT domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage JBD30 (virus) Pseudomonas phage JBD30 (virus) |
| Molecular weight | Theoretical: 33.69498 KDa |
| Sequence | String: MAIITPALIS ALKTSFQKHF QDALATAPST YLQVATVIPS TTASNTYGWL GQFPKLREWI GQRVIKDMAA QGYQITNKLF ESTVGVKRT DIEDDNLGVY GPLMQEMGRA AGAHPDELVF ALLKAGNANL CYDGQNFFDT DHPVYPNVDG TGTATTVSNL F APAADPGA ...String: MAIITPALIS ALKTSFQKHF QDALATAPST YLQVATVIPS TTASNTYGWL GQFPKLREWI GQRVIKDMAA QGYQITNKLF ESTVGVKRT DIEDDNLGVY GPLMQEMGRA AGAHPDELVF ALLKAGNANL CYDGQNFFDT DHPVYPNVDG TGTATTVSNL F APAADPGA AWYLLDTSRS LKPLIYQERM KPSFTSMTKE DDEQVFMADE YRYGVRSRCN VGFGFWQLAA MSTEELNQVN FE KVYDAMR NQKADGGRPL DIRPNLLVVP TTLRSKAKEV VGVQRLANGA DNPNFELVQV LDTAWLN UniProtKB: Bacteriophage Mu GpT domain-containing protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
Details: 10 mM MgSO4, 10 mM NaCl, 50 mM Tris pH 8 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: OTHER / Details: Gatan Solarus II | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: blotting force 0, blotting time 2 s, waiting time 15 s. | ||||||||||||
| Details | phage titer 10^11 PFU |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 12356 / Average exposure time: 2.0 sec. / Average electron dose: 34.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-8rkn: |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)