[English] 日本語
 Yorodumi
Yorodumi- EMDB-19034: Cryo-EM structure of the NADH:ferredoxin oxidoreductase RNF from ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the NADH:ferredoxin oxidoreductase RNF from Azotobacter vinelandii, dithionite reduced | ||||||||||||||||||
 Map data Map data | unsharpened map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | NADH:ferredoxin oxidoreductase / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationTranslocases / electron transport chain / transmembrane transport / FMN binding / 4 iron, 4 sulfur cluster binding / electron transfer activity / metal ion binding / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Azotobacter vinelandii DJ (bacteria) Azotobacter vinelandii DJ (bacteria) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.32 Å | ||||||||||||||||||
 Authors Authors | Zhang L / Einsle O | ||||||||||||||||||
| Funding support | European Union,  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2024 Journal: Nat Chem Biol / Year: 2024Title: Architecture of the RNF1 complex that drives biological nitrogen fixation. Authors: Lin Zhang / Oliver Einsle /  Abstract: Biological nitrogen fixation requires substantial metabolic energy in form of ATP as well as low-potential electrons that must derive from central metabolism. During aerobic growth, the free-living ...Biological nitrogen fixation requires substantial metabolic energy in form of ATP as well as low-potential electrons that must derive from central metabolism. During aerobic growth, the free-living soil diazotroph Azotobacter vinelandii transfers electrons from the key metabolite NADH to the low-potential ferredoxin FdxA that serves as a direct electron donor to the dinitrogenase reductases. This process is mediated by the RNF complex that exploits the proton motive force over the cytoplasmic membrane to lower the midpoint potential of the transferred electron. Here we report the cryogenic electron microscopy structure of the nitrogenase-associated RNF complex of A. vinelandii, a seven-subunit membrane protein assembly that contains four flavin cofactors and six iron-sulfur centers. Its function requires the strict coupling of electron and proton transfer but also involves major conformational changes within the assembly that can be traced with a combination of electron microscopy and modeling. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19034.map.gz emd_19034.map.gz | 75.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19034-v30.xml emd-19034-v30.xml emd-19034.xml emd-19034.xml | 26.9 KB 26.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19034.png emd_19034.png | 81.6 KB | ||
| Masks |  emd_19034_msk_1.map emd_19034_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19034.cif.gz emd-19034.cif.gz | 7.1 KB | ||
| Others |  emd_19034_additional_1.map.gz emd_19034_additional_1.map.gz emd_19034_half_map_1.map.gz emd_19034_half_map_1.map.gz emd_19034_half_map_2.map.gz emd_19034_half_map_2.map.gz | 78 MB 77.7 MB 77.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19034 http://ftp.pdbj.org/pub/emdb/structures/EMD-19034 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19034 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19034 | HTTPS FTP |
-Validation report
| Summary document |  emd_19034_validation.pdf.gz emd_19034_validation.pdf.gz | 909.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19034_full_validation.pdf.gz emd_19034_full_validation.pdf.gz | 909.4 KB | Display | |
| Data in XML |  emd_19034_validation.xml.gz emd_19034_validation.xml.gz | 12.5 KB | Display | |
| Data in CIF |  emd_19034_validation.cif.gz emd_19034_validation.cif.gz | 14.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19034 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19034 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19034 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19034 | HTTPS FTP |
-Related structure data
| Related structure data |  8rbqMC  8ahxC  8rb8C  8rb9C  8rbmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19034.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19034.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
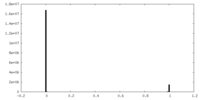
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19034_msk_1.map emd_19034_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
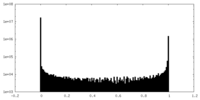
| Density Histograms |
-Additional map: sharpened map
| File | emd_19034_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_19034_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
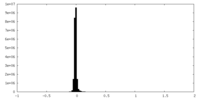
| Density Histograms |
-Half map: half map A
| File | emd_19034_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
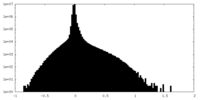
| Density Histograms |
- Sample components
Sample components
+Entire : Heptameric complex NADH:ferredoxin oxidoreductase RNF1
+Supramolecule #1: Heptameric complex NADH:ferredoxin oxidoreductase RNF1
+Macromolecule #1: Ion-translocating oxidoreductase complex subunit A
+Macromolecule #2: Ion-translocating oxidoreductase complex subunit C
+Macromolecule #3: Ion-translocating oxidoreductase complex subunit E
+Macromolecule #4: Ion-translocating oxidoreductase complex subunit G
+Macromolecule #5: Protein RnfH
+Macromolecule #6: Ion-translocating oxidoreductase complex subunit B
+Macromolecule #7: Ion-translocating oxidoreductase complex subunit D
+Macromolecule #8: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #9: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #10: FLAVIN MONONUCLEOTIDE
+Macromolecule #11: IRON/SULFUR CLUSTER
+Macromolecule #12: PHOSPHATIDYLETHANOLAMINE
+Macromolecule #13: RIBOFLAVIN
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 37.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.32 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 165903 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)