[English] 日本語
 Yorodumi
Yorodumi- EMDB-18134: Cryo-EM structure of the DNA polymerase holoenzyme E9-A20-D4 of v... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the DNA polymerase holoenzyme E9-A20-D4 of vaccinia virus | ||||||||||||||||||||||||
 Map data Map data | Sharpened map with b=160 A2, masked. | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | DNA polymerase / holoenzyme / processivity factor / uracil-DNA glycosylase / VIRAL PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationuracil-DNA glycosylase / viral DNA genome replication / uracil DNA N-glycosylase activity / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / DNA-templated DNA replication / DNA recombination / DNA replication / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / hydrolase activity ...uracil-DNA glycosylase / viral DNA genome replication / uracil DNA N-glycosylase activity / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / DNA-templated DNA replication / DNA recombination / DNA replication / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / hydrolase activity / DNA repair / nucleotide binding / DNA binding Similarity search - Function | ||||||||||||||||||||||||
| Biological species |   Vaccinia virus Copenhagen Vaccinia virus Copenhagen | ||||||||||||||||||||||||
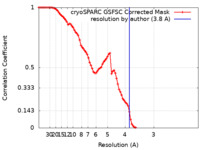
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | ||||||||||||||||||||||||
 Authors Authors | Burmeister WP / Ballandras-Colas A / Boettcher B / Grimm C | ||||||||||||||||||||||||
| Funding support |  France, France,  United States, United States,  Germany, 7 items Germany, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2024 Journal: PLoS Pathog / Year: 2024Title: Structure and flexibility of the DNA polymerase holoenzyme of vaccinia virus. Authors: Wim P Burmeister / Laetitia Boutin / Aurelia C Balestra / Henri Gröger / Allison Ballandras-Colas / Stephanie Hutin / Christian Kraft / Clemens Grimm / Bettina Böttcher / Utz Fischer / ...Authors: Wim P Burmeister / Laetitia Boutin / Aurelia C Balestra / Henri Gröger / Allison Ballandras-Colas / Stephanie Hutin / Christian Kraft / Clemens Grimm / Bettina Böttcher / Utz Fischer / Nicolas Tarbouriech / Frédéric Iseni /   Abstract: The year 2022 was marked by the mpox outbreak caused by the human monkeypox virus (MPXV), which is approximately 98% identical to the vaccinia virus (VACV) at the sequence level with regard to the ...The year 2022 was marked by the mpox outbreak caused by the human monkeypox virus (MPXV), which is approximately 98% identical to the vaccinia virus (VACV) at the sequence level with regard to the proteins involved in DNA replication. We present the production in the baculovirus-insect cell system of the VACV DNA polymerase holoenzyme, which consists of the E9 polymerase in combination with its co-factor, the A20-D4 heterodimer. This led to the 3.8 Å cryo-electron microscopy (cryo-EM) structure of the DNA-free form of the holoenzyme. The model of the holoenzyme was constructed from high-resolution structures of the components of the complex and the A20 structure predicted by AlphaFold 2. The structures do not change in the context of the holoenzyme compared to the previously determined crystal and NMR structures, but the E9 thumb domain became disordered. The E9-A20-D4 structure shows the same compact arrangement with D4 folded back on E9 as observed for the recently solved MPXV holoenzyme structures in the presence and the absence of bound DNA. A conserved interface between E9 and D4 is formed by a cluster of hydrophobic residues. Small-angle X-ray scattering data show that other, more open conformations of E9-A20-D4 without the E9-D4 contact exist in solution using the flexibility of two hinge regions in A20. Biolayer interferometry (BLI) showed that the E9-D4 interaction is indeed weak and transient in the absence of DNA although it is very important, as it has not been possible to obtain viable viruses carrying mutations of key residues within the E9-D4 interface. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18134.map.gz emd_18134.map.gz | 53 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18134-v30.xml emd-18134-v30.xml emd-18134.xml emd-18134.xml | 25.1 KB 25.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18134_fsc.xml emd_18134_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_18134.png emd_18134.png | 205.8 KB | ||
| Filedesc metadata |  emd-18134.cif.gz emd-18134.cif.gz | 7.8 KB | ||
| Others |  emd_18134_half_map_1.map.gz emd_18134_half_map_1.map.gz emd_18134_half_map_2.map.gz emd_18134_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18134 http://ftp.pdbj.org/pub/emdb/structures/EMD-18134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18134 | HTTPS FTP |
-Validation report
| Summary document |  emd_18134_validation.pdf.gz emd_18134_validation.pdf.gz | 640.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18134_full_validation.pdf.gz emd_18134_full_validation.pdf.gz | 640.1 KB | Display | |
| Data in XML |  emd_18134_validation.xml.gz emd_18134_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  emd_18134_validation.cif.gz emd_18134_validation.cif.gz | 21.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18134 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18134 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18134 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18134 | HTTPS FTP |
-Related structure data
| Related structure data |  8q3rMC  8qamC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18134.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18134.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map with b=160 A2, masked. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.064 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: correct hand, from cryosparc NU refinement
| File | emd_18134_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | correct hand, from cryosparc NU refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: correct hand, from cryosparc NU refinement
| File | emd_18134_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | correct hand, from cryosparc NU refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : E9-A20-D4 DNA polymerase holoenzyme
| Entire | Name: E9-A20-D4 DNA polymerase holoenzyme |
|---|---|
| Components |
|
-Supramolecule #1: E9-A20-D4 DNA polymerase holoenzyme
| Supramolecule | Name: E9-A20-D4 DNA polymerase holoenzyme / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: heterotrimer |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 196.825 kDa/nm |
-Macromolecule #1: Uracil-DNA glycosylase
| Macromolecule | Name: Uracil-DNA glycosylase / type: protein_or_peptide / ID: 1 / Details: D4 carries a N-terminal Strep-tag / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vaccinia virus Copenhagen Vaccinia virus Copenhagen |
| Molecular weight | Theoretical: 27.468379 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASWSHPQFE KSGGGGGLVP RGSAMNSVTV SHAPYTITYH DDWEPVMSQL VEFYNEVASW LLRDETSPIP DKFFIQLKQP LRNKRVCVC GIDPYPKDGT GVPFESPNFT KKSIKEIASS ISRLTGVIDY KGYNLNIIDG VIPWNYYLSC KLGETKSHAI Y WDKISKLL ...String: MASWSHPQFE KSGGGGGLVP RGSAMNSVTV SHAPYTITYH DDWEPVMSQL VEFYNEVASW LLRDETSPIP DKFFIQLKQP LRNKRVCVC GIDPYPKDGT GVPFESPNFT KKSIKEIASS ISRLTGVIDY KGYNLNIIDG VIPWNYYLSC KLGETKSHAI Y WDKISKLL LQHITKHVSV LYCLGKTDFS NIRAKLESPV TTIVGYHPAA RDRQFEKDRS FEIINVLLEL DNKAPINWAQ GF IY UniProtKB: Uracil-DNA glycosylase |
-Macromolecule #2: DNA polymerase processivity factor component OPG148
| Macromolecule | Name: DNA polymerase processivity factor component OPG148 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vaccinia virus Copenhagen Vaccinia virus Copenhagen |
| Molecular weight | Theoretical: 49.247031 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MTSSADLTNL KELLSLYKSL RFSDSAAIEK YNSLVEWGTS TYWKIGVQKV ANVETSISDY YDEVKNKPFN IDPGYYIFLP VYFGSVFIY SKGKNMVELG SGNSFQIPDD MRSACNKVLD SDNGIDFLRF VLLNNRWIME DAISKYQSPV NIFKLASEYG L NIPKYLEI ...String: MTSSADLTNL KELLSLYKSL RFSDSAAIEK YNSLVEWGTS TYWKIGVQKV ANVETSISDY YDEVKNKPFN IDPGYYIFLP VYFGSVFIY SKGKNMVELG SGNSFQIPDD MRSACNKVLD SDNGIDFLRF VLLNNRWIME DAISKYQSPV NIFKLASEYG L NIPKYLEI EIEEDTLFDD ELYSIIERSF DDKFPKISIS YIKLGELRRQ VVDFFKFSFM YIESIKVDRI GDNIFIPSVI TK SGKKILV KDVDHLIRSK VREHTFVKVK KKNTFSILYD YDGNGTETRG EVIKRIIDTI GRDYYVNGKY FSKVGSAGLK QLT NKLDIN ECATVDELVD EINKSGTVKR KIKNQSAFDL SRECLGYPEA DFITLVNNMR FKIENCKVVN FNIENTNCLN NPSI ETIYR NFNQFVSIFN VVTDVKKRLF E UniProtKB: DNA polymerase processivity factor component OPG148 |
-Macromolecule #3: DNA polymerase
| Macromolecule | Name: DNA polymerase / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vaccinia virus Copenhagen Vaccinia virus Copenhagen |
| Molecular weight | Theoretical: 120.361484 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSYYHHHHHH DYDIPTTENL YFQGAMDPDV RCINWFESHG ENRFLYLKSR CRNGETVFIR FPHYFYYVVT DEIYQSLSPP PFNARPLGK MRTIDIDETI SYNLDIKDRK CSVADMWLIE EPKKRSIQNA TMDEFLNISW FYISNGISPD GCYSLDEQYL T KINNGCYH ...String: MSYYHHHHHH DYDIPTTENL YFQGAMDPDV RCINWFESHG ENRFLYLKSR CRNGETVFIR FPHYFYYVVT DEIYQSLSPP PFNARPLGK MRTIDIDETI SYNLDIKDRK CSVADMWLIE EPKKRSIQNA TMDEFLNISW FYISNGISPD GCYSLDEQYL T KINNGCYH CDDPRNCFAK KIPRFDIPRS YLFLDIECHF DKKFPSVFIN PISHTSYCYI DLSGKRLLFT LINEEMLTEQ EI QEAVDRG CLRIQSLMEM DYERELVLCS EIVLLRIAKQ LLELTFDYVV TFNGHNFDLR YITNRLELLT GEKIIFRSPD KKE AVHLCI YERNQSSHKG VGGMANTTFH VNNNNGTIFF DLYSFIQKSE KLDSYKLDSI SKNAFSCMGK VLNRGVREMT FIGD DTTDA KGKAAAFAKV LTTGNYVTVD EDIICKVIRK DIWENGFKVV LLCPTLPNDT YKLSFGKDDV DLAQMYKDYN LNIAL DMAR YCIHDACLCQ YLWEYYGVET KTDAGASTYV LPQSMVFEYR ASTVIKGPLL KLLLETKTIL VRSETKQKFP YEGGKV FAP KQKMFSNNVL IFDYNSLYPN VCIFGNLSPE TLVGVVVSTN RLEEEINNQL LLQKYPPPRY ITVHCEPRLP NLISEIA IF DRSIEGTIPR LLRTFLAERA RYKKMLKQAT SSTEKAIYDS MQYTYKIVAN SVYGLMGFRN SALYSYASAK SCTSIGRR M ILYLESVLNG AELSNGMLRF ANPLSNPFYM DDRDINPIVK TSLPIDYRFR FRSVYGDTDS VFTEIDSQDV DKSIEIAKE LERLINNRVL FNNFKIEFEA VYKNLIMQSK KKYTTMKYSA SSNSKSVPER INKGTSETRR DVSKFHKNMI KTYKTRLSEM LSEGRMNSN QVCIDILRSL ETDLRSEFDS RSSPLELFML SRMHHSNYKS ADNPNMYLVT EYNKNNPETI ELGERYYFAY I CPANVPWT KKLVNIKTYE TIIDRSFKLG SDQRIFYEVY FKRLTSEIVN LLDNKVLCIS FFERMFGSKP TFYEA UniProtKB: DNA polymerase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.3 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
Details: 35 mM Tris-HCl, pH 8, 150 mM NaCl, 5 mM EDTA, 3.8 mM desthiobiotin | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.013000000000000001 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 1376 / Average electron dose: 68.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 42000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Details | Phenix real-space refinement | ||||||||||||
| Refinement | Space: REAL / Protocol: OTHER | ||||||||||||
| Output model |  PDB-8q3r: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)