[English] 日本語
 Yorodumi
Yorodumi- EMDB-17730: masked refinement giving rise to better defined protruding densit... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | masked refinement giving rise to better defined protruding densities of the potential macrodomain outside the AUD helical assemblies. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Helical scaffold / Replication complex / Alpha granules / Viral factories / VIRAL PROTEIN | |||||||||
| Biological species |  Chikungunya virus strain S27-African prototype Chikungunya virus strain S27-African prototype | |||||||||
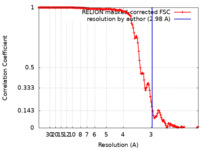
| Method | helical reconstruction / cryo EM / Resolution: 2.98 Å | |||||||||
 Authors Authors | Reguera J / Hons M / Zimberger C / Ptchelkine D / Jones R / Desfosses A | |||||||||
| Funding support |  France, 2 items France, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: The alphavirus nsP3 protein forms helical tubular scaffolds important for viral replication and particle assembly Authors: Reguera J / Hons M / Zimberger C / Ptchelkine D / Jones R / Desfosses A | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17730.map.gz emd_17730.map.gz | 203.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17730-v30.xml emd-17730-v30.xml emd-17730.xml emd-17730.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17730_fsc.xml emd_17730_fsc.xml | 14.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_17730.png emd_17730.png | 119.9 KB | ||
| Filedesc metadata |  emd-17730.cif.gz emd-17730.cif.gz | 5.3 KB | ||
| Others |  emd_17730_additional_1.map.gz emd_17730_additional_1.map.gz emd_17730_additional_2.map.gz emd_17730_additional_2.map.gz emd_17730_half_map_1.map.gz emd_17730_half_map_1.map.gz emd_17730_half_map_2.map.gz emd_17730_half_map_2.map.gz | 5.7 MB 200.2 MB 204.5 MB 204.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17730 http://ftp.pdbj.org/pub/emdb/structures/EMD-17730 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17730 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17730 | HTTPS FTP |
-Validation report
| Summary document |  emd_17730_validation.pdf.gz emd_17730_validation.pdf.gz | 766.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17730_full_validation.pdf.gz emd_17730_full_validation.pdf.gz | 765.6 KB | Display | |
| Data in XML |  emd_17730_validation.xml.gz emd_17730_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  emd_17730_validation.cif.gz emd_17730_validation.cif.gz | 28.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17730 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17730 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17730 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17730 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17730.map.gz / Format: CCP4 / Size: 259.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17730.map.gz / Format: CCP4 / Size: 259.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
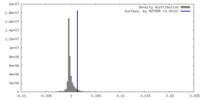
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Class of a mask including 16 protomers Map alligned to EMD-17729
| File | emd_17730_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Class of a mask including 16 protomers Map alligned to EMD-17729 | ||||||||||||
| Projections & Slices |
| ||||||||||||
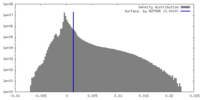
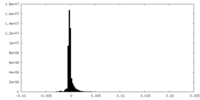
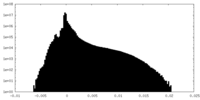
| Density Histograms |
-Additional map: Map alligned to EMD-17729
| File | emd_17730_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map alligned to EMD-17729 | ||||||||||||
| Projections & Slices |
| ||||||||||||
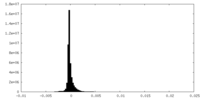
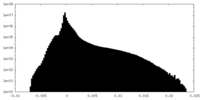
| Density Histograms |
-Half map: #1
| File | emd_17730_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
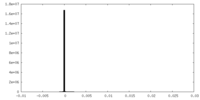
| Density Histograms |
-Half map: #2
| File | emd_17730_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
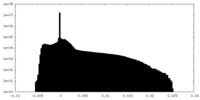
| Density Histograms |
- Sample components
Sample components
-Entire : Helical scaffold assembly of CHIKV nsP3 mediated by its Unique al...
| Entire | Name: Helical scaffold assembly of CHIKV nsP3 mediated by its Unique alphavirus domain. |
|---|---|
| Components |
|
-Supramolecule #1: Helical scaffold assembly of CHIKV nsP3 mediated by its Unique al...
| Supramolecule | Name: Helical scaffold assembly of CHIKV nsP3 mediated by its Unique alphavirus domain. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Chikungunya virus strain S27-African prototype Chikungunya virus strain S27-African prototype |
| Molecular weight | Theoretical: 220.25 kDa/nm |
-Macromolecule #1: Chikungunya virus non structural protein 3
| Macromolecule | Name: Chikungunya virus non structural protein 3 / type: protein_or_peptide / ID: 1 / Details: Zn / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chikungunya virus strain S27-African prototype Chikungunya virus strain S27-African prototype |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: APSYRVKRMD IAKNDEECVV NAANPRGLPG DGVCKAVYKK WPESFKNSAT PVGTAKTVMC GTYPVIHAVG PNFSNYSESE GDRELAAAYR EVAKEVTRLG VNSVAIPLLS TGVYSGGKDR LTQSLNHLFT AMDSTDADVV IYCRDKEWEK KISEAIQMRT QVELLDEHIS ...String: APSYRVKRMD IAKNDEECVV NAANPRGLPG DGVCKAVYKK WPESFKNSAT PVGTAKTVMC GTYPVIHAVG PNFSNYSESE GDRELAAAYR EVAKEVTRLG VNSVAIPLLS TGVYSGGKDR LTQSLNHLFT AMDSTDADVV IYCRDKEWEK KISEAIQMRT QVELLDEHIS IDCDVVRVHP DSSLAGRKGY STTEGALYSY LEGTRFHQTA VDMAEIYTMW PKQTEANEQV CLYALGESIE SIRQKCPVDD ADASSPPKTV PCLCRYAMTP ERVTRLRMNH VTSIIVCSSF PLPKYKIEGV QKVKCSKVML FDHNVPSRVS PREYRPSQES VQEASTTTSL THSQFDLSVD GKILPVPSDL DADAPALEPA LDDGAIHTLP SATGNLAAVS DWVMSTVPVA PPRRRRGRNL TVTCDEREGN ITPMASVRFF RAELCPVVQE TAETRDTAMS LQAPPSTATE LSHPPISFGA PSETFPITFG DFNEGEIESL SSELLTFGDF LPGEVDDLTD SDWSTCSDTD DEL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
| Details | his sample was heterogeneous in lenght |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 18.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)