+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human full length RAD52 undecamer | |||||||||
 Map data Map data | Human full length RAD52 cryo-electron density map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA repair protein / oligomeric structure / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdouble-strand break repair via single-strand annealing / DNA double-strand break processing involved in repair via single-strand annealing / DNA recombinase assembly / mitotic recombination / regulation of nucleotide-excision repair / HDR through MMEJ (alt-NHEJ) / HDR through Single Strand Annealing (SSA) / SUMOylation of DNA damage response and repair proteins / double-strand break repair via homologous recombination / protein-DNA complex ...double-strand break repair via single-strand annealing / DNA double-strand break processing involved in repair via single-strand annealing / DNA recombinase assembly / mitotic recombination / regulation of nucleotide-excision repair / HDR through MMEJ (alt-NHEJ) / HDR through Single Strand Annealing (SSA) / SUMOylation of DNA damage response and repair proteins / double-strand break repair via homologous recombination / protein-DNA complex / double-strand break repair / single-stranded DNA binding / cellular response to oxidative stress / DNA recombination / protein-containing complex / DNA binding / nucleoplasm / identical protein binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
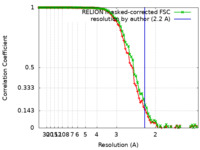
| Method | single particle reconstruction / cryo EM / Resolution: 2.2 Å | |||||||||
 Authors Authors | Marotta R / Balboni B / Girotto S / Cavalli A | |||||||||
| Funding support | European Union,  Italy, 2 items Italy, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2024 Journal: Commun Biol / Year: 2024Title: An integrative structural study of the human full-length RAD52 at 2.2 Å resolution. Authors: Beatrice Balboni / Roberto Marotta / Francesco Rinaldi / Giulia Milordini / Giulia Varignani / Stefania Girotto / Andrea Cavalli /   Abstract: Human RAD52 (RAD52) is a DNA-binding protein involved in many DNA repair mechanisms and genomic stability maintenance. In the last few years, this protein was discovered to be a promising novel ...Human RAD52 (RAD52) is a DNA-binding protein involved in many DNA repair mechanisms and genomic stability maintenance. In the last few years, this protein was discovered to be a promising novel pharmacological target for anticancer strategies. Although the interest in RAD52 has exponentially grown in the previous decade, most information about its structure and mechanism still needs to be elucidated. Here, we report the 2.2 Å resolution cryo-EM reconstruction of the full-length RAD52 (FL-RAD52) protein. This allows us to describe the hydration shell of the N-terminal region of FL-RAD52, which is structured in an undecamer ring. Water molecules coordinate with protein residues to promote stabilization inside and among the protomers and within the inner DNA binding cleft to drive protein-DNA recognition. Additionally, through a multidisciplinary approach involving SEC-SAXS and computational methods, we comprehensively describe the highly flexible and dynamic organization of the C-terminal portion of FL-RAD52. This work discloses unprecedented structural details on the FL-RAD52, which will be critical for characterizing its mechanism of action and inhibitor development, particularly in the context of novel approaches to synthetic lethality and anticancer drug discovery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16089.map.gz emd_16089.map.gz | 27.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16089-v30.xml emd-16089-v30.xml emd-16089.xml emd-16089.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16089_fsc.xml emd_16089_fsc.xml emd_16089_fsc_2.xml emd_16089_fsc_2.xml | 9.6 KB 7.5 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_16089.png emd_16089.png | 34.9 KB | ||
| Filedesc metadata |  emd-16089.cif.gz emd-16089.cif.gz | 6.1 KB | ||
| Others |  emd_16089_half_map_1.map.gz emd_16089_half_map_1.map.gz emd_16089_half_map_2.map.gz emd_16089_half_map_2.map.gz | 27.1 MB 27.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16089 http://ftp.pdbj.org/pub/emdb/structures/EMD-16089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16089 | HTTPS FTP |
-Validation report
| Summary document |  emd_16089_validation.pdf.gz emd_16089_validation.pdf.gz | 787.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16089_full_validation.pdf.gz emd_16089_full_validation.pdf.gz | 786.7 KB | Display | |
| Data in XML |  emd_16089_validation.xml.gz emd_16089_validation.xml.gz | 13.5 KB | Display | |
| Data in CIF |  emd_16089_validation.cif.gz emd_16089_validation.cif.gz | 18.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16089 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16089 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16089 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16089 | HTTPS FTP |
-Related structure data
| Related structure data |  8bjmMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16089.map.gz / Format: CCP4 / Size: 35.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16089.map.gz / Format: CCP4 / Size: 35.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human full length RAD52 cryo-electron density map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.731 Å | ||||||||||||||||||||||||||||||||||||
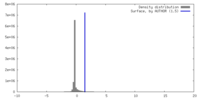
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Human full-length RAD52 half cryo-electron density map 1
| File | emd_16089_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human full-length RAD52 half cryo-electron density map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
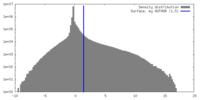
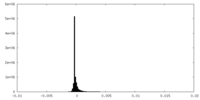
| Density Histograms |
-Half map: Human full-length RAD52 half cryo-electron density map 2
| File | emd_16089_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human full-length RAD52 half cryo-electron density map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
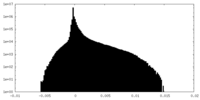
| Density Histograms |
- Sample components
Sample components
-Entire : Human full length RAD52
| Entire | Name: Human full length RAD52 |
|---|---|
| Components |
|
-Supramolecule #1: Human full length RAD52
| Supramolecule | Name: Human full length RAD52 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: N-terminal 6xHis-RAD52 recombinant protein |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 650 KDa |
-Macromolecule #1: DNA repair protein RAD52 homolog
| Macromolecule | Name: DNA repair protein RAD52 homolog / type: protein_or_peptide / ID: 1 / Number of copies: 11 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 48.044637 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHENL YFQGMSGTEE AILGGRDSHP AAGGGSVLCF GQCQYTAEEY QAIQKALRQR LGPEYISSRM AGGGQKVCYI EGHRVINLA NEMFGYNGWA HSITQQNVDF VDLNNGKFYV GVCAFVRVQL KDGSYHEDVG YGVSEGLKSK ALSLEKARKE A VTDGLKRA ...String: MHHHHHHENL YFQGMSGTEE AILGGRDSHP AAGGGSVLCF GQCQYTAEEY QAIQKALRQR LGPEYISSRM AGGGQKVCYI EGHRVINLA NEMFGYNGWA HSITQQNVDF VDLNNGKFYV GVCAFVRVQL KDGSYHEDVG YGVSEGLKSK ALSLEKARKE A VTDGLKRA LRSFGNALGN CILDKDYLRS LNKLPRQLPL EVDLTKAKRQ DLEPSVEEAR YNSCRPNMAL GHPQLQQVTS PS RPSHAVI PADQDCSSRS LSSSAVESEA THQRKLRQKQ LQQQFRERME KQQVRVSTPS AEKSEAAPPA PPVTHSTPVT VSE PLLEKD FLAGVTQELI KTLEDNSEKW AVTPDAGDGV VKPSSRADPA QTSDTLALNN QMVTQNRTPH SVCHQKPQAK SGSW DLQTY SADQRTTGNW ESHRKSQDMK KRKYDPS UniProtKB: DNA repair protein RAD52 homolog |
-Macromolecule #2: water
| Macromolecule | Name: water / type: ligand / ID: 2 / Number of copies: 498 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 318.15 K / Instrument: FEI VITROBOT MARK IV |
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 17400 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)