+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Core divisome complex FtsWIQBL from Pseudomonas aeruginosa | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacterial cell division / peptidoglycan synthesis / membrane protein complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationlipid-linked peptidoglycan transporter activity / cell septum assembly / peptidoglycan glycosyltransferase / peptidoglycan glycosyltransferase activity / cell septum / serine-type D-Ala-D-Ala carboxypeptidase / division septum assembly / serine-type D-Ala-D-Ala carboxypeptidase activity / FtsZ-dependent cytokinesis / cell division site ...lipid-linked peptidoglycan transporter activity / cell septum assembly / peptidoglycan glycosyltransferase / peptidoglycan glycosyltransferase activity / cell septum / serine-type D-Ala-D-Ala carboxypeptidase / division septum assembly / serine-type D-Ala-D-Ala carboxypeptidase activity / FtsZ-dependent cytokinesis / cell division site / penicillin binding / peptidoglycan biosynthetic process / cell wall organization / regulation of cell shape / cell division / proteolysis / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Kaeshammer L / van den Ent F / Jeffery M / Lowe J | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2023 Journal: Nat Microbiol / Year: 2023Title: Cryo-EM structure of the bacterial divisome core complex and antibiotic target FtsWIQBL. Authors: Lisa Käshammer / Fusinita van den Ent / Magnus Jeffery / Nicolas L Jean / Victoria L Hale / Jan Löwe /  Abstract: In most bacteria, cell division relies on the synthesis of new cell wall material by the multiprotein divisome complex. Thus, at the core of the divisome are the transglycosylase FtsW, which ...In most bacteria, cell division relies on the synthesis of new cell wall material by the multiprotein divisome complex. Thus, at the core of the divisome are the transglycosylase FtsW, which synthesises peptidoglycan strands from its substrate Lipid II, and the transpeptidase FtsI that cross-links these strands to form a mesh, shaping and protecting the bacterial cell. The FtsQ-FtsB-FtsL trimeric complex interacts with the FtsWI complex and is involved in regulating its enzymatic activities; however, the structure of this pentameric complex is unknown. Here, we present the cryogenic electron microscopy structure of the FtsWIQBL complex from Pseudomonas aeruginosa at 3.7 Å resolution. Our work reveals intricate structural details, including an extended coiled coil formed by FtsL and FtsB and the periplasmic interaction site between FtsL and FtsI. Our structure explains the consequences of previously reported mutations and we postulate a possible activation mechanism involving a large conformational change in the periplasmic domain. As FtsWIQBL is central to the divisome, our structure is foundational for the design of future experiments elucidating the precise mechanism of bacterial cell division, an important antibiotic target. #2:  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: Divisome core complex in bacterial cell division revealed by cryo-EM Authors: Kashammer L / van den Ent F / Jeffery M / Jean NL / Hale VL / Lowe J | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16042.map.gz emd_16042.map.gz | 78.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16042-v30.xml emd-16042-v30.xml emd-16042.xml emd-16042.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16042.png emd_16042.png | 86.7 KB | ||
| Filedesc metadata |  emd-16042.cif.gz emd-16042.cif.gz | 6.6 KB | ||
| Others |  emd_16042_half_map_1.map.gz emd_16042_half_map_1.map.gz emd_16042_half_map_2.map.gz emd_16042_half_map_2.map.gz | 70.8 MB 70.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16042 http://ftp.pdbj.org/pub/emdb/structures/EMD-16042 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16042 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16042 | HTTPS FTP |
-Validation report
| Summary document |  emd_16042_validation.pdf.gz emd_16042_validation.pdf.gz | 859.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16042_full_validation.pdf.gz emd_16042_full_validation.pdf.gz | 858.7 KB | Display | |
| Data in XML |  emd_16042_validation.xml.gz emd_16042_validation.xml.gz | 13 KB | Display | |
| Data in CIF |  emd_16042_validation.cif.gz emd_16042_validation.cif.gz | 14.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16042 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16042 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16042 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16042 | HTTPS FTP |
-Related structure data
| Related structure data |  8bh1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16042.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16042.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
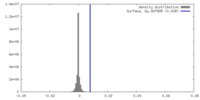
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_16042_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
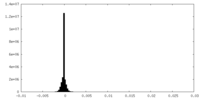
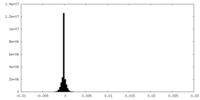
| Density Histograms |
-Half map: #1
| File | emd_16042_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
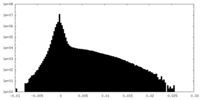
| Density Histograms |
- Sample components
Sample components
-Entire : FtsWIQBL
| Entire | Name: FtsWIQBL |
|---|---|
| Components |
|
-Supramolecule #1: FtsWIQBL
| Supramolecule | Name: FtsWIQBL / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
| Molecular weight | Theoretical: 164 KDa |
-Macromolecule #1: Probable peptidoglycan glycosyltransferase FtsW
| Macromolecule | Name: Probable peptidoglycan glycosyltransferase FtsW / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: peptidoglycan glycosyltransferase |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 48.24143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTAWSHPQFE KGSAGSAAGS GAGWSHPQFE KGLEVLFQGP GGSSMLSVLR PFPSPLLSRH GIDLDFPLLA GCLALLGLGL VMVTSASSE VAAAQSGNPL YFSVRHLIYL VIGLISCGLT MMVPMATWQR WGWKLLLVAF GLLVLVITPG IGREVNGSMR W IGFGLFNI ...String: MTAWSHPQFE KGSAGSAAGS GAGWSHPQFE KGLEVLFQGP GGSSMLSVLR PFPSPLLSRH GIDLDFPLLA GCLALLGLGL VMVTSASSE VAAAQSGNPL YFSVRHLIYL VIGLISCGLT MMVPMATWQR WGWKLLLVAF GLLVLVITPG IGREVNGSMR W IGFGLFNI QPSEIAKVCV VIFMAGYLIR RQQEVRESWM GFFKPFVVLL PMAGLLLREP DFGATVVMMG AAAAMLFLGG VG LFRFGLM VLLAVGAVVL LIQTQPYRMA RLTNFTDPWA DQFGAGYQLS QALIAFGRGG WLGMGLGNSI QKQFYLPEAH TDF VFAVLA EELGIVGALA TVALFVFVSL RALYIGIWAE QAKQFFSAYV AYGLAFLWIG QFLINIGVNV GLLPTKGLTL PFLS YGGSS LVICCACLGM LLRIEWERRT HLGSEEYEFN EEDFADER UniProtKB: Probable peptidoglycan glycosyltransferase FtsW |
-Macromolecule #2: Peptidoglycan D,D-transpeptidase FtsI
| Macromolecule | Name: Peptidoglycan D,D-transpeptidase FtsI / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: serine-type D-Ala-D-Ala carboxypeptidase |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 62.933082 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKLNYFQGAL YPWRFCVIVG LLLAMVGAIV WRIVDLHVID HDFLKGQGDA RSVRHIAIPA HRGLITDRNG EPLAVSTPVT TLWANPKEL MTAKERWPQL AAALGQDTKL FADRIEQNAE REFIYLVRGL TPEQGEGVIA LKVPGVYSIE EFRRFYPAGE V VAHAVGFT ...String: MKLNYFQGAL YPWRFCVIVG LLLAMVGAIV WRIVDLHVID HDFLKGQGDA RSVRHIAIPA HRGLITDRNG EPLAVSTPVT TLWANPKEL MTAKERWPQL AAALGQDTKL FADRIEQNAE REFIYLVRGL TPEQGEGVIA LKVPGVYSIE EFRRFYPAGE V VAHAVGFT DVDDRGREGI ELAFDEWLAG VPGKRQVLKD RRGRVIKDVQ VTKNAKPGKT LALSIDLRLQ YLAHRELRNA LL ENGAKAG SLVIMDVKTG EILAMTNQPT YNPNNRRNLQ PAAMRNRAMI DVFEPGSTVK PFSMSAALAS GRWKPSDIVD VYP GTLQIG RYTIRDVSRN SRQLDLTGIL IKSSNVGISK IAFDIGAESI YSVMQQVGLG QDTGLGFPGE RVGNLPNHRK WPKA ETATL AYGYGLSVTA IQLAHAYAAL ANDGKSVPLS MTRVDRVPDG VQVISPEVAS TVQGMLQQVV EAQGGVFRAQ VPGYH AAGK SGTARKVSVG TKGYRENAYR SLFAGFAPAT DPRIAMVVVI DEPSKAGYFG GLVSAPVFSK VMAGALRLMN VPPDNL PTA TEQQQVNAAP AKGGRG UniProtKB: Peptidoglycan D,D-transpeptidase FtsI |
-Macromolecule #3: Cell division protein FtsQ
| Macromolecule | Name: Cell division protein FtsQ / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 32.290223 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNGVLLRHQQ PGGLGRAPRK PMPRGASRLV AKEPLSVRLP KADFSFLKYL AWPLLLAVLG YGAYRGAEYI LPYADRPIAK VSVEGDLSY ISQRAVQQRI SPYLAASFFT IDLAGMRGQL EQMPWIAHAE VRRVWPDQVV IRLDEQLPIA RWGDEALLNN Q GQAFTPKE ...String: MNGVLLRHQQ PGGLGRAPRK PMPRGASRLV AKEPLSVRLP KADFSFLKYL AWPLLLAVLG YGAYRGAEYI LPYADRPIAK VSVEGDLSY ISQRAVQQRI SPYLAASFFT IDLAGMRGQL EQMPWIAHAE VRRVWPDQVV IRLDEQLPIA RWGDEALLNN Q GQAFTPKE LANYEHLPRL HGPQRAQQQV MQQYQLLSQL LRPLGFSIAR LEMSDRGGWA LTTAQGVEIQ IGRDHVVDKI RR FVSIYDK ALKDQISNIA RIDLRYPNGL AVAWREPVTP ATVATASAVQ UniProtKB: Cell division protein FtsQ |
-Macromolecule #4: Cell division protein FtsL
| Macromolecule | Name: Cell division protein FtsL / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 11.150034 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSRLFVKRLP TGSFLMLLLY IGLLLSAIAV AYSTYWNRQL LNSLYSELSV RDKAQAEWGR LILEQSTWTA HSRIESLAVE QLRMRVPDP AEVRMVAP UniProtKB: Cell division protein FtsL |
-Macromolecule #5: Cell division protein FtsB
| Macromolecule | Name: Cell division protein FtsB / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 12.295922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRLRSPYWLF VVLILALAGL QYRLWVGDGS LAQVRDLQKQ IADQHGENER LLERNRILEA EVAELKKGTE TVEERARHEL GMVKDGETL YQLAKGGSSG GSSHHHHHH UniProtKB: Cell division protein FtsB |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 41.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 136364 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 4.0) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 4.0) |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)