+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | IAPP S20G growth-phase fibril polymorph 3PF-CU | |||||||||
 Map data Map data | CryoEM map for 6-week aged IAPP-S20G 3PF-CU fibrils | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Amyloid / fibril / helical / cross-beta / Amylin / polymorph / islet / PROTEIN FIBRIL / Diabetes | |||||||||
| Function / homology |  Function and homology information Function and homology informationamylin receptor signaling pathway / Calcitonin-like ligand receptors / negative regulation of amyloid fibril formation / negative regulation of bone resorption / eating behavior / positive regulation of protein kinase A signaling / negative regulation of osteoclast differentiation / Regulation of gene expression in beta cells / negative regulation of protein-containing complex assembly / bone resorption ...amylin receptor signaling pathway / Calcitonin-like ligand receptors / negative regulation of amyloid fibril formation / negative regulation of bone resorption / eating behavior / positive regulation of protein kinase A signaling / negative regulation of osteoclast differentiation / Regulation of gene expression in beta cells / negative regulation of protein-containing complex assembly / bone resorption / sensory perception of pain / positive regulation of calcium-mediated signaling / osteoclast differentiation / hormone activity / cell-cell signaling / amyloid-beta binding / G alpha (s) signalling events / positive regulation of MAPK cascade / receptor ligand activity / positive regulation of apoptotic process / Amyloid fiber formation / signaling receptor binding / lipid binding / apoptotic process / signal transduction / extracellular space / extracellular region / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Wilkinson M / Xu Y / Gallardo R / Radford SE / Ranson NA | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Structural evolution of fibril polymorphs during amyloid assembly. Authors: Martin Wilkinson / Yong Xu / Dev Thacker / Alexander I P Taylor / Declan G Fisher / Rodrigo U Gallardo / Sheena E Radford / Neil A Ranson /  Abstract: Cryoelectron microscopy (cryo-EM) has provided unprecedented insights into amyloid fibril structures, including those associated with disease. However, these structures represent the endpoints of ...Cryoelectron microscopy (cryo-EM) has provided unprecedented insights into amyloid fibril structures, including those associated with disease. However, these structures represent the endpoints of long assembly processes, and their relationship to fibrils formed early in assembly is unknown. Consequently, whether different fibril architectures, with potentially different pathological properties, form during assembly remains unknown. Here, we used cryo-EM to determine structures of amyloid fibrils at different times during in vitro fibrillation of a disease-related variant of human islet amyloid polypeptide (IAPP-S20G). Strikingly, the fibrils formed in the lag, growth, and plateau phases have different structures, with new forms appearing and others disappearing as fibrillation proceeds. A time course with wild-type hIAPP also shows fibrils changing with time, suggesting that this is a general property of IAPP amyloid assembly. The observation of transiently populated fibril structures has implications for understanding amyloid assembly mechanisms with potential new insights into amyloid progression in disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15730.map.gz emd_15730.map.gz | 6.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15730-v30.xml emd-15730-v30.xml emd-15730.xml emd-15730.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15730.png emd_15730.png | 64.3 KB | ||
| Filedesc metadata |  emd-15730.cif.gz emd-15730.cif.gz | 5.6 KB | ||
| Others |  emd_15730_half_map_1.map.gz emd_15730_half_map_1.map.gz emd_15730_half_map_2.map.gz emd_15730_half_map_2.map.gz | 96.6 MB 96.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15730 http://ftp.pdbj.org/pub/emdb/structures/EMD-15730 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15730 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15730 | HTTPS FTP |
-Related structure data
| Related structure data |  8az2MC  8awtC  8az0C  8az1C  8az3C  8az4C  8az5C  8az6C  8az7C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15730.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15730.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map for 6-week aged IAPP-S20G 3PF-CU fibrils | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
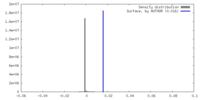
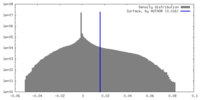
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: halfmap1
| File | emd_15730_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
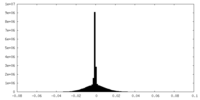
| Density Histograms |
-Half map: halfmap2
| File | emd_15730_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
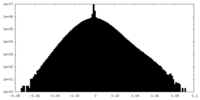
| Density Histograms |
- Sample components
Sample components
-Entire : IAPP S20G growth-phase fibril polymorph 3PF-CU
| Entire | Name: IAPP S20G growth-phase fibril polymorph 3PF-CU |
|---|---|
| Components |
|
-Supramolecule #1: IAPP S20G growth-phase fibril polymorph 3PF-CU
| Supramolecule | Name: IAPP S20G growth-phase fibril polymorph 3PF-CU / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: In vitro fibril growth for 6 weeks at room temperature |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Islet amyloid polypeptide
| Macromolecule | Name: Islet amyloid polypeptide / type: protein_or_peptide / ID: 1 / Details: Synthesised with C-terminal amidation / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.877286 KDa |
| Sequence | String: KCNTATCATQ RLANFLVHSG NNFGAILSST NVGSNTY(NH2) UniProtKB: Islet amyloid polypeptide |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.12 mg/mL |
|---|---|
| Buffer | pH: 6.8 / Component - Concentration: 20.0 mM / Component - Formula: NH3CH3CO2 / Component - Name: ammonium acetate |
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 6s blot. |
| Details | Fibrillation conditions: 30 uM monomeric IAPP-S20G, quiescent at room temp for 6 weeks |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 2350 / Average exposure time: 5.0 sec. / Average electron dose: 39.0 e/Å2 Details: 1204 raw EER frames were collected per image and combined into 30 fractions for processing |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 4.81 Å Applied symmetry - Helical parameters - Δ&Phi: -1.99 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 4.0) / Number images used: 7639 |
|---|---|
| Segment selection | Number selected: 842792 Details: Manually picked a subset of images to train a model for automatic fibril segment picking in crYOLO |
| Startup model | Type of model: INSILICO MODEL Details: Model generated from 2D class averages using relion_helix_inimodel2d |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: RELION (ver. 4.0) |
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 60 / Target criteria: Correlation coefficient | ||||||
| Output model |  PDB-8az2: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)