[English] 日本語
 Yorodumi
Yorodumi- EMDB-14172: CryoEM structure of bacterial transcription intermediate complex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of bacterial transcription intermediate complex mediated by activator protein PspF | |||||||||
 Map data Map data | Local filtered map: mismatch RPi 2nd map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA polymerase / AAA protein / transcription regulation / cryoEM / TRANSCRIPTION | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Ye FZ / Zhang XD | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Mechanisms of DNA opening revealed in AAA+ transcription complex structures. Authors: Fuzhou Ye / Forson Gao / Xiaojiao Liu / Martin Buck / Xiaodong Zhang /  Abstract: Gene transcription is carried out by RNA polymerase (RNAP) and requires the conversion of the initial closed promoter complex, where DNA is double stranded, to a transcription-competent open promoter ...Gene transcription is carried out by RNA polymerase (RNAP) and requires the conversion of the initial closed promoter complex, where DNA is double stranded, to a transcription-competent open promoter complex, where DNA is opened up. In bacteria, RNAP relies on σ factors for its promoter specificities. Using a special form of sigma factor (σ), which forms a stable closed complex and requires its activator that belongs to the AAA+ ATPases (ATPases associated with diverse cellular activities), we obtained cryo-electron microscopy structures of transcription initiation complexes that reveal a previously unidentified process of DNA melting opening. The σ amino terminus threads through the locally opened up DNA and then becomes enclosed by the AAA+ hexameric ring in the activator-bound intermediate complex. Our structures suggest how ATP hydrolysis by the AAA+ activator could remove the σ inhibition while helping to open up DNA, using σ amino-terminal peptide as a pry bar. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14172.map.gz emd_14172.map.gz | 49.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14172-v30.xml emd-14172-v30.xml emd-14172.xml emd-14172.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14172.png emd_14172.png | 157 KB | ||
| Filedesc metadata |  emd-14172.cif.gz emd-14172.cif.gz | 6.4 KB | ||
| Others |  emd_14172_additional_1.map.gz emd_14172_additional_1.map.gz emd_14172_additional_2.map.gz emd_14172_additional_2.map.gz | 47.9 MB 45.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14172 http://ftp.pdbj.org/pub/emdb/structures/EMD-14172 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14172 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14172 | HTTPS FTP |
-Validation report
| Summary document |  emd_14172_validation.pdf.gz emd_14172_validation.pdf.gz | 413.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14172_full_validation.pdf.gz emd_14172_full_validation.pdf.gz | 412.9 KB | Display | |
| Data in XML |  emd_14172_validation.xml.gz emd_14172_validation.xml.gz | 6.1 KB | Display | |
| Data in CIF |  emd_14172_validation.cif.gz emd_14172_validation.cif.gz | 7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14172 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14172 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14172 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14172 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14172.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14172.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local filtered map: mismatch RPi 2nd map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
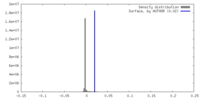
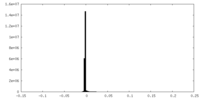
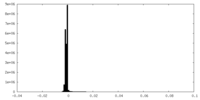
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Focused refinement map: holoenzyme-DNA
| File | emd_14172_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused refinement map: holoenzyme-DNA | ||||||||||||
| Projections & Slices |
| ||||||||||||
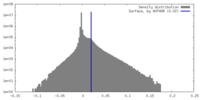
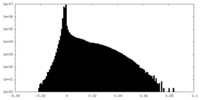
| Density Histograms |
-Additional map: Focused refinement map: PspF-DNA-sigma54
| File | emd_14172_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused refinement map: PspF-DNA-sigma54 | ||||||||||||
| Projections & Slices |
| ||||||||||||
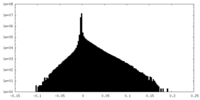
| Density Histograms |
- Sample components
Sample components
-Entire : Bacterial transcription intermediate complex mediated by PspF act...
| Entire | Name: Bacterial transcription intermediate complex mediated by PspF activator protein |
|---|---|
| Components |
|
-Supramolecule #1: Bacterial transcription intermediate complex mediated by PspF act...
| Supramolecule | Name: Bacterial transcription intermediate complex mediated by PspF activator protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 0.71 kDa/nm |
-Macromolecule #1: Bacterial transcription intermediate compelx
| Macromolecule | Name: Bacterial transcription intermediate compelx / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MVYSYTEKKR IRKDFGKRPQ VLDVPYLLSI QLDSFQKFIE QDPEGQYGLE AAFRSVFPIQ SYSGNSELQY VSYRLGEPVF DVQECQIRG VTYSAPLRVK LRLVIYEREA PEGTVKDIKE QEVYMGEIPL MTDNGTFVIN GTERVIVSQL HRSPGVFFDS D KGKTHSSG ...String: MVYSYTEKKR IRKDFGKRPQ VLDVPYLLSI QLDSFQKFIE QDPEGQYGLE AAFRSVFPIQ SYSGNSELQY VSYRLGEPVF DVQECQIRG VTYSAPLRVK LRLVIYEREA PEGTVKDIKE QEVYMGEIPL MTDNGTFVIN GTERVIVSQL HRSPGVFFDS D KGKTHSSG KVLYNARIIP YRGSWLDFEF DPKDNLFVRI DRRRKLPATI ILRALNYTTE QILDLFFEKV IFEIRDNKLQ ME LVPERLR GETASFDIEA NGKVYVEKGR RITARHIRQL EKDDVKLIEV PVEYIAGKVV AKDYIDESTG ELICAANMEL SLD LLAKLS QSGHKRIETL FTNDLDHGPY ISETLRVDPT NDRLSALVEI YRMMRPGEPP TREAAESLFE NLFFSEDRYD LSAV GRMKF NRSLLREEIE GSGILSKDDI IDVMKKLIDI RNGKGEVDDI DHLGNRRIRS VGEMAENQFR VGLVRVERAV KERLS LGDL DTLMPQDMIN AKPISAAVKE FFGSSQLSQF MDQNNPLSEI THKRRISALG PGGLTRERAG FEVRDVHPTH YGRVCP IET PEGPNIGLIN SLSVYAQTNE YGFLETPYRK VTDGVVTDEI HYLSAIEEGN YVIAQANSNL DEEGHFVEDL VTCRSKG ES SLFSRDQVDY MDVSTQQVVS VGASLIPFLE HDDANRALMG ANMQRQAVPT LRADKPLVGT GMERAVAVDS GVTAVAKR G GVVQYVDASR IVIKVNEDEM YPGEAGIDIY NLTKYTRSNQ NTCINQMPCV SLGEPVERGD VLADGPSTDL GELALGQNM RVAFMPWNGY NFEDSILVSE RVVQEDRFTT IHIQELACVS RDTKLGPEEI TADIPNVGEA ALSKLDESGI VYIGAEVTGG DILVGKVTP KGETQLTPEE KLLRAIFGEK ASDVKDSSLR VPNGVSGTVI DVQVFTRDGV EKDKRALEIE EMQLKQAKKD L SEELQILE AGLFSRIRAV LVAGGVEAEK LDKLPRDRWL ELGLTDEEKQ NQLEQLAEQY DELKHEFEKK LEAKRRKITQ GD DLAPGVL KIVKVYLAVK RRIQPGDKMA GRHGNKGVIS KINPIEDMPY DENGTPVDIV LNPLGVPSRM NIGQILETHL GMA AKGIGD KINAMLKQQQ EVAKLREFIQ RAYDLGADVR QKVDLSTFSD EEVMRLAENL RKGMPIATPV FDGAKEAEIK ELLK LGDLP TSGQIRLYDG RTGEQFERPV TVGYMYMLKL NHLVDDKMHA RSTGSYSLVT QQPLGGKAQF GGQRFGEMEV WALEA YGAA YTLQEMLTVK SDDVNGRTKM YKNIVDGNHQ MEPGMPESFN VLLKEIRSLG INIELEDE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 20 mM Tris, 150 mM KCl, 10 mM MgCl2, 8 mM CHAPSO | |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | The complexed was formed in vitro by mixing different components in a certain order to assembly into functional complex. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 14780 / Average exposure time: 4.1 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)