+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-14086 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 S Omicron Spike B.1.1.529 | |||||||||
 Map data Map data | full map for the SARS cov2 omicron spike | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 S Omicron Spike B.1.1.529 / VIRAL PROTEIN | |||||||||
| Function / homology | Fibritin C-terminal / Fibritin C-terminal region / Fibritin / Surface glycoprotein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
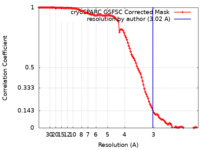
| Method | single particle reconstruction / cryo EM / Resolution: 3.02 Å | |||||||||
 Authors Authors | Ni D / Lau K | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: Structural analysis of the Spike of the Omicron SARS-COV-2 variant by cryo-EM and implications for immune evasion Authors: Ni D / Lau K / Turelli P / Raclot C / Beckert B / Nazarov S / Pojer F / Myasnikov A / Stahlberg H / Trono D | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14086.map.gz emd_14086.map.gz | 82.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14086-v30.xml emd-14086-v30.xml emd-14086.xml emd-14086.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14086_fsc.xml emd_14086_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_14086.png emd_14086.png | 115.3 KB | ||
| Filedesc metadata |  emd-14086.cif.gz emd-14086.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14086 http://ftp.pdbj.org/pub/emdb/structures/EMD-14086 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14086 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14086 | HTTPS FTP |
-Related structure data
| Related structure data |  7qo7MC  7qo9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14086.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14086.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full map for the SARS cov2 omicron spike | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0671 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
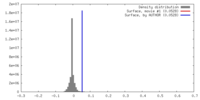
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Spike Trimer
| Entire | Name: Spike Trimer |
|---|---|
| Components |
|
-Supramolecule #1: Spike Trimer
| Supramolecule | Name: Spike Trimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 752 kDa/nm |
-Macromolecule #1: Surface glycoprotein,Fibritin,SARS-CoV-2 S Omicron Spike B.1.1.529
| Macromolecule | Name: Surface glycoprotein,Fibritin,SARS-CoV-2 S Omicron Spike B.1.1.529 type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 142.558094 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVY FASIEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL DHKNNKSWME SEFRVYSSAN N CTFEYVSQ ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVY FASIEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL DHKNNKSWME SEFRVYSSAN N CTFEYVSQ PFLMDLEGKQ GNFKNLREFV FKNIDGYFKI YSKHTPIIVR EPEDLPQGFS ALEPLVDLPI GINITRFQTL LA LHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPT ESIVRF PNITNLCPFD EVFNATRFAS VYAWNRKRIS NCVADYSVLY NLAPFFTFKC YGVSPTKLND LCFTNVYADS FVIR GDEVR QIAPGQTGNI ADYNYKLPDD FTGCVIAWNS NKLDSKVSGN YNYLYRLFRK SNLKPFERDI STEIYQAGNK PCNGV AGFN CYFPLRSYSF RPTYGVGHQP YRVVVLSFEL LHAPATVCGP KKSTNLVKNK CVNFNFNGLK GTGVLTESNK KFLPFQ QFG RDIADTTDAV RDPQTLEILD ITPCSFGGVS VITPGTNTSN QVAVLYQGVN CTEVPVAIHA DQLTPTWRVY STGSNVF QT RAGCLIGAEY VNNSYECDIP IGAGICASYQ TQTKSHGSAS SVASQSIIAY TMSLGAENSV AYSNNSIAIP TNFTISVT T EILPVSMTKT SVDCTMYICG DSTECSNLLL QYGSFCTQLK RALTGIAVEQ DKNTQEVFAQ VKQIYKTPPI KYFGGFNFS QILPDPSKPS KRSFIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFKGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGA ALQIPFAMQM AYRFNGIGVT QNVLYENQKL IANQFNSAIG KIQDSLSSTA SALGKLQDVV NHNAQALNTL V KQLSSKFG AISSVLNDIF SRLDPPEAEV QIDRLITGRL QSLQTYVTQQ LIRAAEIRAS ANLAATKMSE CVLGQSKRVD FC GKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVS GNCDVV IGIVNNTVYD PLQPELDSFK EELDKYFKNH TSPDVDLGDI SGINASVVNI QKEIDRLNEV AKNLNESLID LQEL GKYEQ GSGYIPEAPR DGQAYVRKDG EWVLLSTFLG RSLEVLFQGP GHHHHHHHHS AWSHPQFEKG GGSGGGGSGG SAWSH PQFE K UniProtKB: Surface glycoprotein, Fibritin |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 14 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.35 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: PBS |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)