[English] 日本語
 Yorodumi
Yorodumi- EMDB-13543: HsPepT1 bound to Ala-Phe in the outward facing open conformation -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13543 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | HsPepT1 bound to Ala-Phe in the outward facing open conformation | |||||||||
 Map data Map data | post processed map in Phenix used for refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HsPepT1 / PepT1 / Peptide transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationproton-dependent oligopeptide secondary active transmembrane transporter activity / tripeptide import across plasma membrane / Proton/oligopeptide cotransporters / tripeptide transmembrane transporter activity / dipeptide import across plasma membrane / peptide:proton symporter activity / dipeptide transmembrane transporter activity / peptide transport / brush border / monoatomic ion transport ...proton-dependent oligopeptide secondary active transmembrane transporter activity / tripeptide import across plasma membrane / Proton/oligopeptide cotransporters / tripeptide transmembrane transporter activity / dipeptide import across plasma membrane / peptide:proton symporter activity / dipeptide transmembrane transporter activity / peptide transport / brush border / monoatomic ion transport / protein transport / apical plasma membrane / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
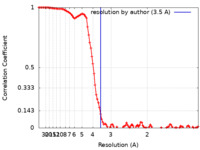
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Killer M / Wald J | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Structural snapshots of human PepT1 and PepT2 reveal mechanistic insights into substrate and drug transport across epithelial membranes. Authors: Maxime Killer / Jiri Wald / Joanna Pieprzyk / Thomas C Marlovits / Christian Löw /  Abstract: The uptake of peptides in mammals plays a crucial role in nutrition and inflammatory diseases. This process is mediated by promiscuous transporters of the solute carrier family 15, which form part of ...The uptake of peptides in mammals plays a crucial role in nutrition and inflammatory diseases. This process is mediated by promiscuous transporters of the solute carrier family 15, which form part of the major facilitator superfamily. Besides the uptake of short peptides, peptide transporter 1 (PepT1) is a highly abundant drug transporter in the intestine and represents a major route for oral drug delivery. PepT2 also allows renal drug reabsorption from ultrafiltration and brain-to-blood efflux of neurotoxic compounds. Here, we present cryogenic electron microscopy (cryo-EM) structures of human PepT1 and PepT2 captured in four different states throughout the transport cycle. The structures reveal the architecture of human peptide transporters and provide mechanistic insights into substrate recognition and conformational transitions during transport. This may support future drug design efforts to increase the bioavailability of different drugs in the human body. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13543.map.gz emd_13543.map.gz | 90.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13543-v30.xml emd-13543-v30.xml emd-13543.xml emd-13543.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13543_fsc.xml emd_13543_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_13543.png emd_13543.png | 87.8 KB | ||
| Masks |  emd_13543_msk_1.map emd_13543_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13543.cif.gz emd-13543.cif.gz | 5.7 KB | ||
| Others |  emd_13543_additional_1.map.gz emd_13543_additional_1.map.gz emd_13543_half_map_1.map.gz emd_13543_half_map_1.map.gz emd_13543_half_map_2.map.gz emd_13543_half_map_2.map.gz | 90.9 MB 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13543 http://ftp.pdbj.org/pub/emdb/structures/EMD-13543 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13543 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13543 | HTTPS FTP |
-Validation report
| Summary document |  emd_13543_validation.pdf.gz emd_13543_validation.pdf.gz | 814.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13543_full_validation.pdf.gz emd_13543_full_validation.pdf.gz | 814.3 KB | Display | |
| Data in XML |  emd_13543_validation.xml.gz emd_13543_validation.xml.gz | 18 KB | Display | |
| Data in CIF |  emd_13543_validation.cif.gz emd_13543_validation.cif.gz | 22.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13543 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13543 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13543 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13543 | HTTPS FTP |
-Related structure data
| Related structure data |  7pmxMC  7pmwC  7pmyC  7pn1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13543.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13543.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post processed map in Phenix used for refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.67 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
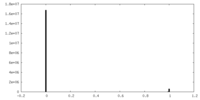
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13543_msk_1.map emd_13543_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
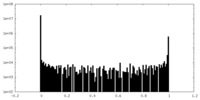
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: post processed map in deepEMhancer used for illustration...
| File | emd_13543_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post processed map in deepEMhancer used for illustration only (tightTarget model) | ||||||||||||
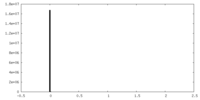
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_13543_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
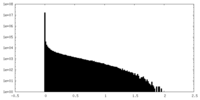
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13543_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human PepT1
| Entire | Name: Human PepT1 |
|---|---|
| Components |
|
-Supramolecule #1: Human PepT1
| Supramolecule | Name: Human PepT1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: ALA-PHE
| Supramolecule | Name: ALA-PHE / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 15 member 1
| Macromolecule | Name: Solute carrier family 15 member 1 / type: protein_or_peptide / ID: 1 / Details: HsPepT1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 78.872422 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGMSKSHSFF GYPLSIFFIV VNEFCERFSY YGMRAILILY FTNFISWDDN LSTAIYHTFV ALCYLTPILG ALIADSWLGK FKTIVSLSI VYTIGQAVTS VSSINDLTDH NHDGTPDSLP VHVVLSLIGL ALIALGTGGI KPCVSAFGGD QFEEGQEKQR N RFFSIFYL ...String: MGMSKSHSFF GYPLSIFFIV VNEFCERFSY YGMRAILILY FTNFISWDDN LSTAIYHTFV ALCYLTPILG ALIADSWLGK FKTIVSLSI VYTIGQAVTS VSSINDLTDH NHDGTPDSLP VHVVLSLIGL ALIALGTGGI KPCVSAFGGD QFEEGQEKQR N RFFSIFYL AINAGSLLST IITPMLRVQQ CGIHSKQACY PLAFGVPAAL MAVALIVFVL GSGMYKKFKP QGNIMGKVAK CI GFAIKNR FRHRSKAFPK REHWLDWAKE KYDERLISQI KMVTRVMFLY IPLPMFWALF DQQGSRWTLQ ATTMSGKIGA LEI QPDQMQ TVNAILIVIM VPIFDAVLYP LIAKCGFNFT SLKKMAVGMV LASMAFVVAA IVQVEIDKTL PVFPKGNEVQ IKVL NIGNN TMNISLPGEM VTLGPMSQTN AFMTFDVNKL TRINISSPGS PVTAVTDDFK QGQRHTLLVW APNHYQVVKD GLNQK PEKG ENGIRFVNTF NELITITMSG KVYANISSYN ASTYQFFPSG IKGFTISSTE IPPQCQPNFN TFYLEFGSAY TYIVQR KND SCPEVKVFED ISANTVNMAL QIPQYFLLTC GEVVFSVTGL EFSYSQAPSN MKSVLQAGWL LTVAVGNIIV LIVAGAG QF SKQWAEYILF AALLLVVCVI FAIMARFYTY INPAEIEAQF DEDEKKNRLE KSNPYFMSGA NSQKQM UniProtKB: Solute carrier family 15 member 1 |
-Macromolecule #2: ALA-PHE
| Macromolecule | Name: ALA-PHE / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 236.267 Da |
| Sequence | String: AF |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: PROPANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)