[English] 日本語
 Yorodumi
Yorodumi- EMDB-13335: Structure of Adenylyl cyclase 9 in complex with Gs protein alpha ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13335 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Adenylyl cyclase 9 in complex with Gs protein alpha subunit and MANT-GTP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein / adenylyl cyclase / signalling transduction. / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationAdenylate cyclase activating pathway / Adenylate cyclase inhibitory pathway / sensory perception of chemical stimulus / PKA activation / adenylate cyclase / Hedgehog 'off' state / cAMP biosynthetic process / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / adenylate cyclase activity ...Adenylate cyclase activating pathway / Adenylate cyclase inhibitory pathway / sensory perception of chemical stimulus / PKA activation / adenylate cyclase / Hedgehog 'off' state / cAMP biosynthetic process / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / adenylate cyclase activity / beta-2 adrenergic receptor binding / G alpha (z) signalling events / D1 dopamine receptor binding / adenylate cyclase-activating adrenergic receptor signaling pathway / insulin-like growth factor receptor binding / ionotropic glutamate receptor binding / adenylate cyclase activator activity / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / adenylate cyclase-activating dopamine receptor signaling pathway / heterotrimeric G-protein complex / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / in utero embryonic development / intracellular signal transduction / ciliary basal body / GTPase activity / GTP binding / ATP binding / metal ion binding / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
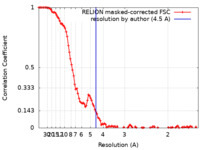
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Qi C / Korkhov VM | |||||||||
| Funding support | 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis of adenylyl cyclase 9 activation. Authors: Chao Qi / Pia Lavriha / Ved Mehta / Basavraj Khanppnavar / Inayathulla Mohammed / Yong Li / Michalis Lazaratos / Jonas V Schaefer / Birgit Dreier / Andreas Plückthun / Ana-Nicoleta Bondar / ...Authors: Chao Qi / Pia Lavriha / Ved Mehta / Basavraj Khanppnavar / Inayathulla Mohammed / Yong Li / Michalis Lazaratos / Jonas V Schaefer / Birgit Dreier / Andreas Plückthun / Ana-Nicoleta Bondar / Carmen W Dessauer / Volodymyr M Korkhov /    Abstract: Adenylyl cyclase 9 (AC9) is a membrane-bound enzyme that converts ATP into cAMP. The enzyme is weakly activated by forskolin, fully activated by the G protein Gαs subunit and is autoinhibited by the ...Adenylyl cyclase 9 (AC9) is a membrane-bound enzyme that converts ATP into cAMP. The enzyme is weakly activated by forskolin, fully activated by the G protein Gαs subunit and is autoinhibited by the AC9 C-terminus. Although our recent structural studies of the AC9-Gαs complex provided the framework for understanding AC9 autoinhibition, the conformational changes that AC9 undergoes in response to activator binding remains poorly understood. Here, we present the cryo-EM structures of AC9 in several distinct states: (i) AC9 bound to a nucleotide inhibitor MANT-GTP, (ii) bound to an artificial activator (DARPin C4) and MANT-GTP, (iii) bound to DARPin C4 and a nucleotide analogue ATPαS, (iv) bound to Gαs and MANT-GTP. The artificial activator DARPin C4 partially activates AC9 by binding at a site that overlaps with the Gαs binding site. Together with the previously observed occluded and forskolin-bound conformations, structural comparisons of AC9 in the four conformations described here show that secondary structure rearrangements in the region surrounding the forskolin binding site are essential for AC9 activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13335.map.gz emd_13335.map.gz | 16.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13335-v30.xml emd-13335-v30.xml emd-13335.xml emd-13335.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13335_fsc.xml emd_13335_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_13335.png emd_13335.png | 43.4 KB | ||
| Filedesc metadata |  emd-13335.cif.gz emd-13335.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13335 http://ftp.pdbj.org/pub/emdb/structures/EMD-13335 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13335 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13335 | HTTPS FTP |
-Related structure data
| Related structure data |  7pdeMC  7pd4C  7pd8C  7pddC  7pdfC  7pdgC  7pdhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13335.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13335.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Adenylyl cyclase 9 bound to MANT-GTP
| Entire | Name: Adenylyl cyclase 9 bound to MANT-GTP |
|---|---|
| Components |
|
-Supramolecule #1: Adenylyl cyclase 9 bound to MANT-GTP
| Supramolecule | Name: Adenylyl cyclase 9 bound to MANT-GTP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Molecular weight | Theoretical: 146 kDa/nm |
-Supramolecule #2: Adenylate cyclase 9
| Supramolecule | Name: Adenylate cyclase 9 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Supramolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Adenylate cyclase 9
| Macromolecule | Name: Adenylate cyclase 9 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 151.142875 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASPPHQQLL QHHSTEVSCD SSGDSNSVRV RINPKQPSSN SHPKHCKYSI SSSCSSSGDS GGVPRRMGAG GRLRRRKKLP QLFERASSR WWDPKFDSVN LEEACMERCF PQTQRRFRYA LFYIGFACLL WSIYFGVHMK SKLIVMVAPA LCFLVVCVGF F LFTFTKLY ...String: MASPPHQQLL QHHSTEVSCD SSGDSNSVRV RINPKQPSSN SHPKHCKYSI SSSCSSSGDS GGVPRRMGAG GRLRRRKKLP QLFERASSR WWDPKFDSVN LEEACMERCF PQTQRRFRYA LFYIGFACLL WSIYFGVHMK SKLIVMVAPA LCFLVVCVGF F LFTFTKLY ARHYVWTSLV LTLLVFALTL AAQFQVLTPL SGRVDNFNHT RAARPTDTCL SQVGSFSMCI EVLFLLYTVM HL PLYLSLI LGVAYSVLFE TFGYHFQDEA CFASPGAEAL HWELLSRALL HLCIHAIGIH LFIMSQVRSR STFLKVGQSI MHG KDLEVE KALKERMIHS VMPRIIADDL MKQGDEESEN SVKRHATSSP KNRKKKSSIQ KAPIAFRPFK MQQIEEVSIL FADI VGFTK MSANKSAHAL VGLLNDLFGR FDRLCEETKC EKISTLGDCY YCVAGCPEPR ADHAYCCIEM GLGMIRAIEQ FCQEK KEMV NMRVGVHTGT VLCGILGMRR FKFDVWSNDV NLANLMEQLG VAGKVHISEA TAKYLDDRYE MEDGKVTERL GQSVVA DQL KGLKTYLIAG QRAKESHCSC SEALLSGFEV LDGSRVSSGP RGQGTASPGS VSDLAQTVKT FDNLKTCPSC GITFTPK PE AGAEGGAVQN GCQEEPKNSA KASGGPSSKT QNGLLSPPPE EKLTNSQTSL CEILQEKGRW AGVSLDQSAL LPLRFKNI R EKTDAHFVDV IKEDSLMKDY FFKPPINQFS LNFLDPELER AYRTSYQEEV VKSSPVRTFA SATFSSLLDV LLSTTVFLI LSITCFLRYG AASTPPPPAA LAVFGAALLL EILSLVVSVR MVFFLEDVMT CTKRLLEWIA GWLPRHFIGA ILVSLPALAV YSHVTSEFE TNIHSTMFTG SAVLTAVVQY CNFCQLSSWM RSSLATVVGA GPLLLLLYVS LCPDSSTVIS HLDAVQNFSS T RKLCNASL PHDGRSPASL IGQEVILVFF LLLLLVWFLN REFEVSYRLH YHGDVEADLH RTKIQSMRDQ ADWLLRNIIP YH VAEQLKV SQTYSKNHDS GGVIFASIVN FSEFYEENYE GGKECYRVLN ELIGDFDELL SKPDYSSIEK IKTIGATYMA ASG LNATQC RDGSHPQEHL QILFEFAKEM MRVVDDFNNN MLWFNFKLRV GFNHGPLTAG VIGTTKLLYD IWGDTVNIAS RMDT TGVEC RIQVSEESYR VLSKMGYEFD YRGTVNVKGK GQMKTYLYPK CTDSGLVPQH QLSISPDIRV QVDGSIGRSP TDEIA SLVP SVQNPDQVPP GSENNAQTRD AHPSAKRPWK EPVRAEERCR FGKAIEKSDC EEVGMEEANE LTKLNVSERA UniProtKB: Adenylate cyclase type 9 |
-Macromolecule #2: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.003801 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKSTIVKQM RILHVNGFNG GEGGEEDPNA AKSNSDGEK ATKVQDIKNN LKEAIETIVA AMSNLVPPVE LANPENQFRV DYILSVMNVP DFDFPPEFYE HAKALWEDEG V RACYERSN ...String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKSTIVKQM RILHVNGFNG GEGGEEDPNA AKSNSDGEK ATKVQDIKNN LKEAIETIVA AMSNLVPPVE LANPENQFRV DYILSVMNVP DFDFPPEFYE HAKALWEDEG V RACYERSN EYQLIDCAQY FLDKIDVIKQ DDYVPSDQDL LRCRVLTSGI FETKFQVDKV NFHMFDVGGQ RDERRKWIQC FN DVTAIIF VVASSSYNMV IREDNQTNRL QEALNLFKSI WNNRWLRTIS VILFLNKQDL LAEKVLAGKS KIEDYFPEFA RYT TPEDAT PEPGEDPRVT RAKYFIRDEF LRISTASGDG RHYCYPHFTC AVDTENIRRV FNDCRDIIQR MHLRQYELLG GHHH HHHHH UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #3: 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE
| Macromolecule | Name: 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: GSP |
|---|---|
| Molecular weight | Theoretical: 539.246 Da |
| Chemical component information |  ChemComp-GSP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7pde: |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)