+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-12979 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex | ||||||||||||||||||
 マップデータ マップデータ | Structure of the TELO2-TTI1-TTI2 complex and its function in TOR recruitment to the R2TP chaperone | ||||||||||||||||||
 試料 試料 |
| ||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報: / positive regulation of DNA damage checkpoint / TTT Hsp90 cochaperone complex / promoter-enhancer loop anchoring activity / regulation of DNA strand elongation / positive regulation of telomere maintenance in response to DNA damage / establishment of protein localization to chromatin / R2TP complex / TORC2 complex / TORC1 complex ...: / positive regulation of DNA damage checkpoint / TTT Hsp90 cochaperone complex / promoter-enhancer loop anchoring activity / regulation of DNA strand elongation / positive regulation of telomere maintenance in response to DNA damage / establishment of protein localization to chromatin / R2TP complex / TORC2 complex / TORC1 complex / dynein axonemal particle / Swr1 complex / RPAP3/R2TP/prefoldin-like complex / regulation of TOR signaling / regulation of double-strand break repair / positive regulation of telomerase RNA localization to Cajal body / Ino80 complex / protein folding chaperone complex / box C/D snoRNP assembly / regulation of chromosome organization / NuA4 histone acetyltransferase complex / telomeric DNA binding / regulation of DNA replication / TFIID-class transcription factor complex binding / regulation of embryonic development / MLL1 complex / Telomere Extension By Telomerase / positive regulation of double-strand break repair via homologous recombination / telomere maintenance via telomerase / RNA polymerase II core promoter sequence-specific DNA binding / regulation of DNA repair / Deposition of new CENPA-containing nucleosomes at the centromere / DNA helicase activity / positive regulation of DNA repair / TBP-class protein binding / telomere maintenance / cellular response to estradiol stimulus / Formation of the beta-catenin:TCF transactivating complex / DNA Damage Recognition in GG-NER / Hsp90 protein binding / euchromatin / negative regulation of canonical Wnt signaling pathway / chromatin DNA binding / ADP binding / beta-catenin binding / nuclear matrix / transcription corepressor activity / UCH proteinases / cellular response to UV / nucleosome / unfolded protein binding / positive regulation of canonical Wnt signaling pathway / protein folding / HATs acetylate histones / ATPase binding / spermatogenesis / regulation of apoptotic process / DNA recombination / DNA helicase / chromosome, telomeric region / molecular adaptor activity / transcription coactivator activity / nuclear body / protein stabilization / Ub-specific processing proteases / regulation of cell cycle / chromatin remodeling / ribonucleoprotein complex / cadherin binding / cell cycle / RNA polymerase II cis-regulatory region sequence-specific DNA binding / cell division / DNA repair / centrosome / protein-containing complex binding / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / protein kinase binding / positive regulation of DNA-templated transcription / protein homodimerization activity / ATP hydrolysis activity / positive regulation of transcription by RNA polymerase II / extracellular exosome / nucleoplasm / ATP binding / identical protein binding / membrane / nucleus / cytoplasm / cytosol 類似検索 - 分子機能 | ||||||||||||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | ||||||||||||||||||
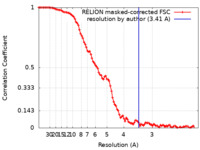
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.41 Å | ||||||||||||||||||
 データ登録者 データ登録者 | Pal M / Llorca O / Pearl L | ||||||||||||||||||
| 資金援助 |  英国, 英国,  スペイン, 5件 スペイン, 5件
| ||||||||||||||||||
 引用 引用 |  ジャーナル: Cell Rep / 年: 2021 ジャーナル: Cell Rep / 年: 2021タイトル: Structure of the TELO2-TTI1-TTI2 complex and its function in TOR recruitment to the R2TP chaperone. 著者: Mohinder Pal / Hugo Muñoz-Hernandez / Dennis Bjorklund / Lihong Zhou / Gianluca Degliesposti / J Mark Skehel / Emma L Hesketh / Rebecca F Thompson / Laurence H Pearl / Oscar Llorca / Chrisostomos Prodromou /   要旨: The R2TP (RUVBL1-RUVBL2-RPAP3-PIH1D1) complex, in collaboration with heat shock protein 90 (HSP90), functions as a chaperone for the assembly and stability of protein complexes, including RNA ...The R2TP (RUVBL1-RUVBL2-RPAP3-PIH1D1) complex, in collaboration with heat shock protein 90 (HSP90), functions as a chaperone for the assembly and stability of protein complexes, including RNA polymerases, small nuclear ribonucleoprotein particles (snRNPs), and phosphatidylinositol 3-kinase (PI3K)-like kinases (PIKKs) such as TOR and SMG1. PIKK stabilization depends on an additional complex of TELO2, TTI1, and TTI2 (TTT), whose structure and function are poorly understood. The cryoelectron microscopy (cryo-EM) structure of the human R2TP-TTT complex, together with biochemical experiments, reveals the mechanism of TOR recruitment to the R2TP-TTT chaperone. The HEAT-repeat TTT complex binds the kinase domain of TOR, without blocking its activity, and delivers TOR to the R2TP chaperone. In addition, TTT regulates the R2TP chaperone by inhibiting RUVBL1-RUVBL2 ATPase activity and by modulating the conformation and interactions of the PIH1D1 and RPAP3 components of R2TP. Taken together, our results show how TTT couples the recruitment of TOR to R2TP with the regulation of this chaperone system. | ||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_12979.map.gz emd_12979.map.gz | 78.4 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-12979-v30.xml emd-12979-v30.xml emd-12979.xml emd-12979.xml | 45.7 KB 45.7 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_12979_fsc.xml emd_12979_fsc.xml | 10 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_12979.png emd_12979.png | 96.3 KB | ||
| その他 |  emd_12979_additional_1.map.gz emd_12979_additional_1.map.gz emd_12979_additional_2.map.gz emd_12979_additional_2.map.gz emd_12979_additional_3.map.gz emd_12979_additional_3.map.gz emd_12979_additional_4.map.gz emd_12979_additional_4.map.gz emd_12979_additional_5.map.gz emd_12979_additional_5.map.gz emd_12979_additional_6.map.gz emd_12979_additional_6.map.gz emd_12979_half_map_1.map.gz emd_12979_half_map_1.map.gz emd_12979_half_map_2.map.gz emd_12979_half_map_2.map.gz | 74.7 MB 74.4 MB 65.3 MB 65.4 MB 65.4 MB 50.4 MB 65.4 MB 65.3 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12979 http://ftp.pdbj.org/pub/emdb/structures/EMD-12979 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12979 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12979 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_12979_validation.pdf.gz emd_12979_validation.pdf.gz | 480 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_12979_full_validation.pdf.gz emd_12979_full_validation.pdf.gz | 479.6 KB | 表示 | |
| XML形式データ |  emd_12979_validation.xml.gz emd_12979_validation.xml.gz | 16.9 KB | 表示 | |
| CIF形式データ |  emd_12979_validation.cif.gz emd_12979_validation.cif.gz | 21.7 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12979 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12979 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12979 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12979 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_12979.map.gz / 形式: CCP4 / 大きさ: 83.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_12979.map.gz / 形式: CCP4 / 大きさ: 83.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Structure of the TELO2-TTI1-TTI2 complex and its function in TOR recruitment to the R2TP chaperone | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
-追加マップ: Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex (dataset1)...
| ファイル | emd_12979_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex (dataset1) | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
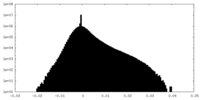
| 密度ヒストグラム |
-追加マップ: Cryo-EM structure of the TELO2-TTI1-TTI2 complex
| ファイル | emd_12979_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Cryo-EM structure of the TELO2-TTI1-TTI2 complex | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
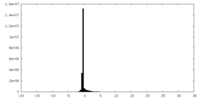
| 密度ヒストグラム |
-追加マップ: Cryo-EM structure of the TELO2-TTI1-TTI2 complex
| ファイル | emd_12979_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Cryo-EM structure of the TELO2-TTI1-TTI2 complex | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
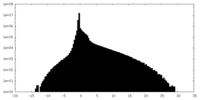
| 密度ヒストグラム |
-追加マップ: Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex (dataset1 halfmap1)...
| ファイル | emd_12979_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex (dataset1 halfmap1) | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
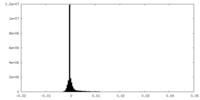
| 密度ヒストグラム |
-追加マップ: Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex (dataset1 halfmap2)...
| ファイル | emd_12979_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex (dataset1 halfmap2) | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-追加マップ: Composite map of the R2-TTT structure
| ファイル | emd_12979_additional_6.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Composite map of the R2-TTT structure | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex (dataset2)...
| ファイル | emd_12979_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex (dataset2) | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex (dataset2)...
| ファイル | emd_12979_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex (dataset2) | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex
| 全体 | 名称: TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex |
|---|---|
| 要素 |
|
-超分子 #1: TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex
| 超分子 | 名称: TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#5 |
|---|---|
| 分子量 | 実験値: 580 KDa |
-分子 #1: RuvB-like 1
| 分子 | 名称: RuvB-like 1 / タイプ: protein_or_peptide / ID: 1 / 詳細: ADP / コピー数: 3 / 光学異性体: LEVO / EC番号: DNA helicase |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 50.296914 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MKIEEVKSTT KTQRIASHSH VKGLGLDESG LAKQAASGLV GQENAREACG VIVELIKSKK MAGRAVLLAG PPGTGKTALA LAIAQELGS KVPFCPMVGS EVYSTEIKKT EVLMENFRRA IGLRIKETKE VYEGEVTELT PCETENPMGG YGKTISHVII G LKTAKGTK ...文字列: MKIEEVKSTT KTQRIASHSH VKGLGLDESG LAKQAASGLV GQENAREACG VIVELIKSKK MAGRAVLLAG PPGTGKTALA LAIAQELGS KVPFCPMVGS EVYSTEIKKT EVLMENFRRA IGLRIKETKE VYEGEVTELT PCETENPMGG YGKTISHVII G LKTAKGTK QLKLDPSIFE SLQKERVEAG DVIYIEANSG AVKRQGRCDT YATEFDLEAE EYVPLPKGDV HKKKEIIQDV TL HDLDVAN ARPQGGQDIL SMMGQLMKPK KTEITDKLRG EINKVVNKYI DQGIAELVPG VLFVDEVHML DIECFTYLHR ALE SSIAPI VIFASNRGNC VIRGTEDITS PHGIPLDLLD RVMIIRTMLY TPQEMKQIIK IRAQTEGINI SEEALNHLGE IGTK TTLRY SVQLLTPANL LAKINGKDSI EKEHVEEISE LFYDAKSSAK ILADQQDKYM K |
-分子 #2: RuvB-like 2
| 分子 | 名称: RuvB-like 2 / タイプ: protein_or_peptide / ID: 2 / 詳細: ADP / コピー数: 3 / 光学異性体: LEVO / EC番号: DNA helicase |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 51.222465 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MATVTATTKV PEIRDVTRIE RIGAHSHIRG LGLDDALEPR QASQGMVGQL AARRAAGVVL EMIREGKIAG RAVLIAGQPG TGKTAIAMG MAQALGPDTP FTAIAGSEIF SLEMSKTEAL TQAFRRSIGV RIKEETEIIE GEVVEIQIDR PATGTGSKVG K LTLKTTEM ...文字列: MATVTATTKV PEIRDVTRIE RIGAHSHIRG LGLDDALEPR QASQGMVGQL AARRAAGVVL EMIREGKIAG RAVLIAGQPG TGKTAIAMG MAQALGPDTP FTAIAGSEIF SLEMSKTEAL TQAFRRSIGV RIKEETEIIE GEVVEIQIDR PATGTGSKVG K LTLKTTEM ETIYDLGTKM IESLTKDKVQ AGDVITIDKA TGKISKLGRS FTRARDYDAM GSQTKFVQCP DGELQKRKEV VH TVSLHEI DVINSRTQGF LALFSGDTGE IKSEVREQIN AKVAEWREEG KAEIIPGVLF IDEVHMLDIE SFSFLNRALE SDM APVLIM ATNRGITRIR GTSYQSPHGI PIDLLDRLLI VSTTPYSEKD TKQILRIRCE EEDVEMSEDA YTVLTRIGLE TSLR YAIQL ITAASLVCRK RKGTEVQVDD IKRVYSLFLD ESRSTQYMKE YQDAFLFNEL KGETMDTS |
-分子 #3: TELO2-interacting protein 1 homolog,TELO2-interacting protein 1 h...
| 分子 | 名称: TELO2-interacting protein 1 homolog,TELO2-interacting protein 1 homolog,TTI1 タイプ: protein_or_peptide / ID: 3 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 176.844344 KDa |
| 組換発現 | 生物種:  Spodoptera aff. frugiperda 1 BOLD-2017 (蝶・蛾) Spodoptera aff. frugiperda 1 BOLD-2017 (蝶・蛾) |
| 配列 | 文字列: MAVFDTPEEA FGVLRPVCVQ LTKTQTVENV EHLQTRLQAV SDSALQELQQ YILFPLRFTL KTPGPKRERL IQSVVECLTF VLSSTCVKE QELLQELFSE LSACLYSPSS QKPAAVSEEL KLAVIQGLST LMHSAYGDII LTFYEPSILP RLGFAVSLLL G LAEQEKSK ...文字列: MAVFDTPEEA FGVLRPVCVQ LTKTQTVENV EHLQTRLQAV SDSALQELQQ YILFPLRFTL KTPGPKRERL IQSVVECLTF VLSSTCVKE QELLQELFSE LSACLYSPSS QKPAAVSEEL KLAVIQGLST LMHSAYGDII LTFYEPSILP RLGFAVSLLL G LAEQEKSK QIKIAALKCL QVLLLQCDCQ DHPRSLDELE QKQLGDLFAS FLPGISTALT RLITGDFKQG HSIVVSSLKI FY KTVSFIM ADEQLKRISK VQAKPAVEHR VAELMVYREA DWVKKTGDKL TILIKKIIEC VSVHPHWKVR LELVELVEDL LLK CSQSLV ECAGPLLKAL VGLVNDESPE IQAQCNKVLR HFADQKVVVG NKALADILSE SLHSLATSLP RLMNSQDDQG KFST LSLLL GYLKLLGPKI NFVLNSVAHL QRLSKALIQV LELDVADIKI VEERRWNSDD LNASPKTSAT QPWNRIQRRY FRFFT DERI FMLLRQVCQL LGYYGNLYLL VDHFMELYHQ SVVYRKQAAM ILNELVTGAA GLEVEDLHEK HIKTNPEELR EIVTSI LEE YTSQENWYLV TCLETEEMGE ELMMEHPGLQ AITSGEHTCQ VTSFLAFSKP SPTICSMNSN IWQICIQLEG IGQFAYA LG KDFCLLLMSA LYPVLEKAGD QTLLISQVAT STMMDVCRAC GYDSLQHLIN QNSDYLVNGI SLNLRHLALH PHTPKVLE V MLRNSDANLL PLVADVVQDV LATLDQFYDK RAASFVSVLH ALMAALAQWF PDTGNLGHLQ EQSLGEEGSH LNQRPAALE KSTTTAEDIE QFLLNYLKEK DVADGNVSDF DNEEEEQSVP PKVDENDTRP DVEPPLPLQI QIAMDVMERC IHLLSDKNLQ IRLKVLDVL DLCVVVLQSH KNQLLPLAHQ AWPSLVHRLT RDAPLAVLRA FKVLRTLGSK CGDFLRSRFC KDVLPKLAGS L VTQAPISA RAGPVYSHTL AFKLQLAVLQ GLGPLCERLD LGEGDLNKVA DACLIYLSVK QPVKLQEAAR SVFLHLMKVD PD STWFLLN ELYCPVQFTP PHPSLHPVQL HGASGQQNPY TTNVLQLLKE LQ(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)G(UNK)G(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)G(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)G (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)G(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)G(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-分子 #4: TELO2-interacting protein 2,TELO2-interacting protein 2,TTI2
| 分子 | 名称: TELO2-interacting protein 2,TELO2-interacting protein 2,TTI2 タイプ: protein_or_peptide / ID: 4 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 95.633781 KDa |
| 組換発現 | 生物種:  Spodoptera aff. frugiperda 1 BOLD-2017 (蝶・蛾) Spodoptera aff. frugiperda 1 BOLD-2017 (蝶・蛾) |
| 配列 | 文字列: MELDSALEAP SQEDSNLSEE LSHSAFGQAF SKILHCLARP EARRGNVKDA VLKDLGDLIE ATEFDRLFEG TGARLRGMPE TLGQVAKAL EKYAAPSKEE EGGGDGHSEA AEKAAQVGLL FLKLLGKVET AKNSLVGPAW QTGLHHLAGP VYIFAITHSL E QPWTTPRS ...文字列: MELDSALEAP SQEDSNLSEE LSHSAFGQAF SKILHCLARP EARRGNVKDA VLKDLGDLIE ATEFDRLFEG TGARLRGMPE TLGQVAKAL EKYAAPSKEE EGGGDGHSEA AEKAAQVGLL FLKLLGKVET AKNSLVGPAW QTGLHHLAGP VYIFAITHSL E QPWTTPRS REVAREVLTS LLQVTECGSV AGFLHGENED EKGRLSVILG LLKPDLYKES WKNNPAIKHV FSWTLQQVTR PW LSQHLER VLPASLVISD DYQTENKILG VHCLHHIVLN VPAADLLQYN RAQVLYHAIS NHLYTPEHHL IQAVLLCLLD LFP ILEKTL HWKGDGARPT THCDEVLRLI LTHMEPEHRL LLRRTYARNL PAFVNRLGIL TVRHLKRLER VIIGYLEVYD GPEE EARLK ILETLKLLMQ HTWPRVSCRL VVLLKALLKL ICDVARDPNL TPESVKSALL QEATDCLILL DRCSQGRVKG LLAKI PQSC EDRKVVNYIR KVQQVSEGAP YNGT(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)G(UNK)G(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)G(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)G (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)G (UNK)(UNK)G (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-分子 #5: Telomere length regulation protein TEL2 homolog
| 分子 | 名称: Telomere length regulation protein TEL2 homolog / タイプ: protein_or_peptide / ID: 5 / 詳細: LGEMEPPALPREKEEFASAHF / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 53.424195 KDa |
| 組換発現 | 生物種:  Spodoptera aff. frugiperda 1 BOLD-2017 (蝶・蛾) Spodoptera aff. frugiperda 1 BOLD-2017 (蝶・蛾) |
| 配列 | 文字列: MEPAPSEVRL AVREAIHALS SSEDGGHIFC TLESLKRYLG EMEPPALPRE (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)KEEFASAHF SPVLRCLASR LSPAWLELLP HGRLE(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) ...文字列: MEPAPSEVRL AVREAIHALS SSEDGGHIFC TLESLKRYLG EMEPPALPRE (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)KEEFASAHF SPVLRCLASR LSPAWLELLP HGRLE(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)ELWASF FLEGPADQAF LVLMETIEGA AGPSFRLMKM ARLLARFLRE GRLAVLMEAQ CRQQTQPGF ILLRETLLGK VV(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)ALPDHLGNR LQQENLAEFF PQNYFRLLGE EVVRVLQAVV DSLQGGLDSS VSFVSQ VLG KACVHGRQQE ILGVLVPRLA ALTQGSYLHQ RVCWRLVEQV PDRAMEAVLT GLVEAALGPE VLSRLLGNLV VKNKKAQ FV MTQKLLFLQS RLTTPMLQSL LGHLAMDSQR RPLLLQVLKE LLETWGSSSA IRHTPLPQQR HVSKAVLICL AQLGEPEL R DSRDELLASM MAGVKCRLDS SLPPVRRLGM IVAEVVSARI HPEGPPLKFQ YEEDELSLEL LALASPQPAG DGASEAGT |
-分子 #6: ADENOSINE-5'-DIPHOSPHATE
| 分子 | 名称: ADENOSINE-5'-DIPHOSPHATE / タイプ: ligand / ID: 6 / コピー数: 5 / 式: ADP |
|---|---|
| 分子量 | 理論値: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7.5 構成要素:
詳細: Solutions were made fresh for protein purification | |||||||||
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: COPPER / メッシュ: 300 / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 雰囲気: AIR | |||||||||
| 凍結 | 凍結剤: ETHANE-PROPANE / チャンバー内湿度: 90 % / チャンバー内温度: 287.15 K / 装置: LEICA PLUNGER / 詳細: 3sec. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 温度 | 最低: 90.0 K / 最高: 100.0 K |
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / デジタル化 - サイズ - 横: 1034 pixel / デジタル化 - サイズ - 縦: 1034 pixel / デジタル化 - サンプリング間隔: 5.0 µm / デジタル化 - 画像ごとのフレーム数: 1-40 / 撮影したグリッド数: 10 / 実像数: 6000 / 平均露光時間: 9.0 sec. / 平均電子線量: 60.17 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 70.0 µm / 最大 デフォーカス(補正後): 3.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最小 デフォーカス(公称値): 1.2 µm / 倍率(公称値): 130000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー

















 Z
Z Y
Y X
X