+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12675 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | GLIC, pentameric ligand gated ion channel, pH 3 State 2 | |||||||||
 Map data Map data | 3D Refined map for pH 3 data State 2 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsodium channel activity / potassium channel activity / transmembrane transporter complex / extracellular ligand-gated monoatomic ion channel activity / transmembrane signaling receptor activity / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Gloeobacter violaceus (bacteria) Gloeobacter violaceus (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM | |||||||||
 Authors Authors | Rovsnik U / Zhuang Y / Forsberg BO / Carroni M / Yvonnesdotter L / Howard RJ / Lindahl E | |||||||||
| Funding support |  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Life Sci Alliance / Year: 2021 Journal: Life Sci Alliance / Year: 2021Title: Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations. Authors: Urška Rovšnik / Yuxuan Zhuang / Björn O Forsberg / Marta Carroni / Linnea Yvonnesdotter / Rebecca J Howard / Erik Lindahl /   Abstract: Ligand-gated ion channels are critical mediators of electrochemical signal transduction across evolution. Biophysical and pharmacological characterization of these receptor proteins relies on high- ...Ligand-gated ion channels are critical mediators of electrochemical signal transduction across evolution. Biophysical and pharmacological characterization of these receptor proteins relies on high-quality structures in multiple, subtly distinct functional states. However, structural data in this family remain limited, particularly for resting and intermediate states on the activation pathway. Here, we report cryo-electron microscopy (cryo-EM) structures of the proton-activated ligand-gated ion channel (GLIC) under three pH conditions. Decreased pH was associated with improved resolution and side chain rearrangements at the subunit/domain interface, particularly involving functionally important residues in the β1-β2 and M2-M3 loops. Molecular dynamics simulations substantiated flexibility in the closed-channel extracellular domains relative to the transmembrane ones and supported electrostatic remodeling around E35 and E243 in proton-induced gating. Exploration of secondary cryo-EM classes further indicated a low-pH population with an expanded pore. These results allow us to define distinct protonation and activation steps in pH-stimulated conformational cycling in GLIC, including interfacial rearrangements largely conserved in the pentameric channel family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12675.map.gz emd_12675.map.gz | 49.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12675-v30.xml emd-12675-v30.xml emd-12675.xml emd-12675.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_12675.png emd_12675.png | 34.2 KB | ||
| Masks |  emd_12675_msk_1.map emd_12675_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_12675_additional_1.map.gz emd_12675_additional_1.map.gz emd_12675_half_map_1.map.gz emd_12675_half_map_1.map.gz emd_12675_half_map_2.map.gz emd_12675_half_map_2.map.gz | 9.3 MB 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12675 http://ftp.pdbj.org/pub/emdb/structures/EMD-12675 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12675 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12675 | HTTPS FTP |
-Validation report
| Summary document |  emd_12675_validation.pdf.gz emd_12675_validation.pdf.gz | 464 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12675_full_validation.pdf.gz emd_12675_full_validation.pdf.gz | 463.6 KB | Display | |
| Data in XML |  emd_12675_validation.xml.gz emd_12675_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_12675_validation.cif.gz emd_12675_validation.cif.gz | 14.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12675 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12675 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12675 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12675 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12675.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12675.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D Refined map for pH 3 data State 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
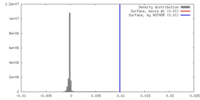
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12675_msk_1.map emd_12675_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
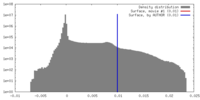
| Density Histograms |
-Additional map: Postprocessed map for pH 3 data State 2
| File | emd_12675_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map for pH 3 data State 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
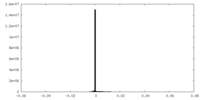
| Density Histograms |
-Half map: Half map 2 for pH 3 data state 2
| File | emd_12675_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 for pH 3 data state 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 for pH 3 data state 2
| File | emd_12675_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 for pH 3 data state 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pentameric ion channel
| Entire | Name: Pentameric ion channel |
|---|---|
| Components |
|
-Supramolecule #1: Pentameric ion channel
| Supramolecule | Name: Pentameric ion channel / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Gloeobacter violaceus (bacteria) Gloeobacter violaceus (bacteria) |
| Recombinant expression | Organism:  |
-Macromolecule #1: Proton-gated ion channel
| Macromolecule | Name: Proton-gated ion channel / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Sequence | String: AQDMVSPPPP IADEPLTVNT GIYLIECYSL DDKAETFKVN AFLSLSWKDR RLAFDPVRSG VRVKTYEPEA IWIPEIRFVN VENARDADVV DISVSPDGTV QYLERFSARV LSPLDFRRYP FDSQTLHIYL IVRSVDTRNI VLAVDLEKVG KNDDVFLTGW DIESFTAVVK ...String: AQDMVSPPPP IADEPLTVNT GIYLIECYSL DDKAETFKVN AFLSLSWKDR RLAFDPVRSG VRVKTYEPEA IWIPEIRFVN VENARDADVV DISVSPDGTV QYLERFSARV LSPLDFRRYP FDSQTLHIYL IVRSVDTRNI VLAVDLEKVG KNDDVFLTGW DIESFTAVVK PANFALEDRL ESKLDYQLRI SRQYFSYIPN IILPMLFILF ISWTAFWSTS YEANVTLVVS TLIAHIAFNI LVETNLPKTP YMTYTGAIIF MIYLFYFVAV IEVTVQHYLK VESQPARAAS ITRASRIAFP VVFLLANIIL AFLFFGF |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 3 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number images used: 23000 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)