[English] 日本語
 Yorodumi
Yorodumi- EMDB-12158: CryoEM structure of Mycobacterium tuberculosis UMP Kinase (UMPK) ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12158 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Mycobacterium tuberculosis UMP Kinase (UMPK) in complex with UDP and UTP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nucleotide metabolism / UMP kinase / allosteric regulation / antibacterial target / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationUMP kinase / UMP kinase activity / 'de novo' CTP biosynthetic process / ATP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.85 Å | |||||||||
 Authors Authors | Bous J / Trapani S | |||||||||
| Funding support | European Union,  France, 2 items France, 2 items
| |||||||||
 Citation Citation |  Journal: FEBS J / Year: 2022 Journal: FEBS J / Year: 2022Title: Structural basis for the allosteric inhibition of UMP kinase from Gram-positive bacteria, a promising antibacterial target. Authors: Patrick Walter / Ariel Mechaly / Julien Bous / Ahmed Haouz / Patrick England / Joséphine Lai-Kee-Him / Aurélie Ancelin / Sylviane Hoos / Bruno Baron / Stefano Trapani / Patrick Bron / ...Authors: Patrick Walter / Ariel Mechaly / Julien Bous / Ahmed Haouz / Patrick England / Joséphine Lai-Kee-Him / Aurélie Ancelin / Sylviane Hoos / Bruno Baron / Stefano Trapani / Patrick Bron / Gilles Labesse / Hélène Munier-Lehmann /  Abstract: Tuberculosis claims significantly more than one million lives each year. A feasible way to face the issue of drug resistance is the development of new antibiotics. Bacterial uridine 5'-monophosphate ...Tuberculosis claims significantly more than one million lives each year. A feasible way to face the issue of drug resistance is the development of new antibiotics. Bacterial uridine 5'-monophosphate (UMP) kinase is a promising target for novel antibiotic discovery as it is essential for bacterial survival and has no counterpart in human cells. The UMP kinase from M. tuberculosis is also a model of particular interest for allosteric regulation with two effectors, GTP (positive) and UTP (negative). In this study, using X-ray crystallography and cryo-electron microscopy, we report for the first time a detailed description of the negative effector UTP-binding site of a typical Gram-positive behaving UMP kinase. Comparison between this snapshot of low affinity for Mg-ATP with our previous 3D-structure of the GTP-bound complex of high affinity for Mg-ATP led to a better understanding of the cooperative mechanism and the allosteric regulation of UMP kinase. Thermal shift assay and circular dichroism experiments corroborate our model of an inhibition by UTP linked to higher flexibility of the Mg-ATP-binding domain. These new structural insights provide valuable knowledge for future drug discovery strategies targeting bacterial UMP kinases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12158.map.gz emd_12158.map.gz | 62.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12158-v30.xml emd-12158-v30.xml emd-12158.xml emd-12158.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12158_fsc.xml emd_12158_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_12158.png emd_12158.png | 113.4 KB | ||
| Masks |  emd_12158_msk_1.map emd_12158_msk_1.map | 67 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12158.cif.gz emd-12158.cif.gz | 6 KB | ||
| Others |  emd_12158_half_map_1.map.gz emd_12158_half_map_1.map.gz emd_12158_half_map_2.map.gz emd_12158_half_map_2.map.gz | 51 MB 50.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12158 http://ftp.pdbj.org/pub/emdb/structures/EMD-12158 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12158 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12158 | HTTPS FTP |
-Validation report
| Summary document |  emd_12158_validation.pdf.gz emd_12158_validation.pdf.gz | 981.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12158_full_validation.pdf.gz emd_12158_full_validation.pdf.gz | 981.4 KB | Display | |
| Data in XML |  emd_12158_validation.xml.gz emd_12158_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_12158_validation.cif.gz emd_12158_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12158 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12158 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12158 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12158 | HTTPS FTP |
-Related structure data
| Related structure data |  7besMC  7bixC  7bl7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12158.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12158.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.052 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
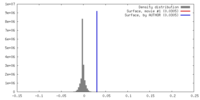
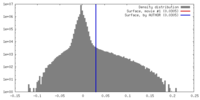
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12158_msk_1.map emd_12158_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
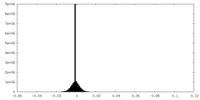
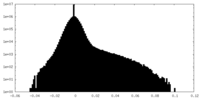
| Density Histograms |
-Half map: #1
| File | emd_12158_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_12158_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : UMPK-UDP-UTP
| Entire | Name: UMPK-UDP-UTP |
|---|---|
| Components |
|
-Supramolecule #1: UMPK-UDP-UTP
| Supramolecule | Name: UMPK-UDP-UTP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 177.558 KDa |
-Macromolecule #1: Uridylate kinase
| Macromolecule | Name: Uridylate kinase / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: UMP kinase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.625893 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MTEPDVAGAP ASKPEPASTG AASAAQLSGY SRVLLKLGGE MFGGGQVGLD PDVVAQVARQ IADVVRGGV QIAVVIGGGN FFRGAQLQQL GMERTRSDYM GMLGTVMNSL ALQDFLEKEG IVTRVQTAIT MGQVAEPYLP L RAVRHLEK ...String: MGSSHHHHHH SSGLVPRGSH MTEPDVAGAP ASKPEPASTG AASAAQLSGY SRVLLKLGGE MFGGGQVGLD PDVVAQVARQ IADVVRGGV QIAVVIGGGN FFRGAQLQQL GMERTRSDYM GMLGTVMNSL ALQDFLEKEG IVTRVQTAIT MGQVAEPYLP L RAVRHLEK GRVVIFGAGM GLPYFSTDTT AAQRALEIGA DVVLMAKAVD GVFAEDPRVN PEAELLTAVS HREVLDRGLR VA DATAFSL CMDNGMPILV FNLLTDGNIA RAVRGEKIGT LVTT UniProtKB: Uridylate kinase |
-Macromolecule #2: URIDINE-5'-DIPHOSPHATE
| Macromolecule | Name: URIDINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 3 / Formula: UDP |
|---|---|
| Molecular weight | Theoretical: 404.161 Da |
| Chemical component information |  ChemComp-UDP: |
-Macromolecule #3: URIDINE 5'-TRIPHOSPHATE
| Macromolecule | Name: URIDINE 5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: UTP |
|---|---|
| Molecular weight | Theoretical: 484.141 Da |
| Chemical component information |  ChemComp-UTP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 / Component:
| ||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 53.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Overall B value: 87.61 |
|---|---|
| Output model |  PDB-7bes: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)