+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11835 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Candida albicans gamma-Tubulin Small Complex | |||||||||
 Map data Map data | gamma-Tubulin Small Complex density map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | gamma-Tubulin Small Complex / Cytoskeleton / Microtubule nucleation / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationinner plaque of spindle pole body / microtubule nucleation by spindle pole body / outer plaque of spindle pole body / gamma-tubulin small complex / gamma-tubulin complex / microtubule nucleation / spindle pole body / gamma-tubulin binding / positive regulation of cytoplasmic translation / spindle assembly ...inner plaque of spindle pole body / microtubule nucleation by spindle pole body / outer plaque of spindle pole body / gamma-tubulin small complex / gamma-tubulin complex / microtubule nucleation / spindle pole body / gamma-tubulin binding / positive regulation of cytoplasmic translation / spindle assembly / cytoplasmic microtubule organization / mitotic spindle organization / meiotic cell cycle / structural constituent of cytoskeleton / spindle pole / mitotic cell cycle / microtubule / GTP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Candida albicans (yeast) Candida albicans (yeast) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Zupa E / Pfeffer S | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: The cryo-EM structure of a γ-TuSC elucidates architecture and regulation of minimal microtubule nucleation systems. Authors: Erik Zupa / Anjun Zheng / Annett Neuner / Martin Würtz / Peng Liu / Anna Böhler / Elmar Schiebel / Stefan Pfeffer /  Abstract: The nucleation of microtubules from αβ-tubulin subunits is mediated by γ-tubulin complexes, which vary in composition across organisms. Aiming to understand how de novo microtubule formation is ...The nucleation of microtubules from αβ-tubulin subunits is mediated by γ-tubulin complexes, which vary in composition across organisms. Aiming to understand how de novo microtubule formation is achieved and regulated by a minimal microtubule nucleation system, we here determined the cryo-electron microscopy structure of the heterotetrameric γ-tubulin small complex (γ-TuSC) from C. albicans at near-atomic resolution. Compared to the vertebrate γ-tubulin ring complex (γ-TuRC), we observed a vastly remodeled interface between the SPC/GCP-γ-tubulin spokes, which stabilizes the complex and defines the γ-tubulin arrangement. The relative positioning of γ-tubulin subunits indicates that a conformational rearrangement of the complex is required for microtubule nucleation activity, which follows opposing directionality as predicted for the vertebrate γ-TuRC. Collectively, our data suggest that the assembly and regulation mechanisms of γ-tubulin complexes fundamentally differ between the microtubule nucleation systems in lower and higher eukaryotes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11835.map.gz emd_11835.map.gz | 39.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11835-v30.xml emd-11835-v30.xml emd-11835.xml emd-11835.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11835.png emd_11835.png | 139.9 KB | ||
| Filedesc metadata |  emd-11835.cif.gz emd-11835.cif.gz | 7.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11835 http://ftp.pdbj.org/pub/emdb/structures/EMD-11835 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11835 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11835 | HTTPS FTP |
-Related structure data
| Related structure data |  7anzMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11835.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11835.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
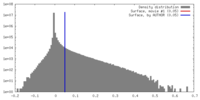
| Annotation | gamma-Tubulin Small Complex density map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : gamma-Tubulin Small Complex
| Entire | Name: gamma-Tubulin Small Complex |
|---|---|
| Components |
|
-Supramolecule #1: gamma-Tubulin Small Complex
| Supramolecule | Name: gamma-Tubulin Small Complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Candida albicans gamma-Tubulin Small Complex expressed and purified from SF21 cells |
|---|---|
| Source (natural) | Organism:  Candida albicans (yeast) Candida albicans (yeast) |
| Molecular weight | Theoretical: 306.7 KDa |
-Macromolecule #1: Tubulin gamma chain
| Macromolecule | Name: Tubulin gamma chain / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candida albicans (yeast) Candida albicans (yeast) |
| Molecular weight | Theoretical: 56.532543 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPGETITLQV GQCGNQVGLQ YWQQLATEHG IQSDGSSTPY PKDINDLQLQ ELNNSGSSPQ SYPQQTKPNG KYRNDHPELF FTLSDSNTY TPRSILIDME PSVIAKSTSA LPMFNPRNVH LSNQGNGAAN NWINGYKYGT EEEETLLNLI DREVDKCDNL S NFQLFHSV ...String: MPGETITLQV GQCGNQVGLQ YWQQLATEHG IQSDGSSTPY PKDINDLQLQ ELNNSGSSPQ SYPQQTKPNG KYRNDHPELF FTLSDSNTY TPRSILIDME PSVIAKSTSA LPMFNPRNVH LSNQGNGAAN NWINGYKYGT EEEETLLNLI DREVDKCDNL S NFQLFHSV AGGTGSGVGS KMLEVISDRY GHKKLLNTFS IFPSNEDTSD VVVQPYNTIL TLKRLIDYSD ATFVFHNDSL NR IENILFN NNSNIQHDDN DLFLGANKLI ALVSASVSNP LRFPGYMYSS MESIVSNLIP TPDLKFLTSS IAPFSTQKHN YLN EYDMLL ELSNDRYKTN RVGGDTSYIS MLNYLIGYNL DQREIRKGIL KSQQRISFVP WVARSVLVVH GKKSPYLKNT NLEG IQVTN NTSMIDVFTK ILKQFDLLIK RKAYLNRYYS SVEEENEVME MFNESRESVK SIIDEYKACK EITYLDDDDE DDLED GDGG GGGNGNGYNN IDDADMGI UniProtKB: Tubulin gamma chain |
-Macromolecule #2: Spindle pole body component
| Macromolecule | Name: Spindle pole body component / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candida albicans (yeast) Candida albicans (yeast) |
| Molecular weight | Theoretical: 101.660453 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNTFSSPPNV IREYNDSTYQ SPLNSQFHQS PFLQTQSPDY VSLREEEDDN NDKNLDIMSS CIVDSVIYKS QKIAGPLLSQ ISNLNIQQA LIIRELLFTL LGHEGHYIQY SKRYDPTSQI SRIEGPDYKI AKNLDISLKV ITKKLVKFGK FYSGLKSFIQ V FDNNKFGK ...String: MNTFSSPPNV IREYNDSTYQ SPLNSQFHQS PFLQTQSPDY VSLREEEDDN NDKNLDIMSS CIVDSVIYKS QKIAGPLLSQ ISNLNIQQA LIIRELLFTL LGHEGHYIQY SKRYDPTSQI SRIEGPDYKI AKNLDISLKV ITKKLVKFGK FYSGLKSFIQ V FDNNKFGK IVQKFCSEVR KFLSSYQQVL INVEHEFKFN KNFNLNMLDS LLHQEISNEM THLYQIGIEI SRITEERQKM SQ AEIMGNF EPTTLANTSM NGINSEPNLY YGKFDCCKGG LLLQVIQERM VYYKGDPTSL DFLTQLFDIV SSDYIGMLNQ WLL EGVIND PFDEFMIREK RVPDSFMEIF QSKSEYYWNE LFLIKIDGLL NQFQNSTIQS KILNTGKYLN IFKRCTGLHN FESL KEKLT TITSLAAPDL ELKIDEFYHR ANKMLMKLLF DGYNFPSVVN IFQRLFLFAD SFQIDNFIDS TFSELKRGKL KISVS RLQK QYDDIFKEKI ENKVGVRPSV YDVLKKNQKL SVTSESLYKV VEELMEKNSD YLISDNNLRG IFHRVASLRD DSRLTI SST ADSATENVKD EPTITSVDLT IPLPFPLNLV LNQQLSYQYE IMFKLLINIK FISKYNSSNW QEMNYSKIWT NSHFNSS VK KWILRCRVLH SRICSFIHEL ENYIVHDVIE HNFEEIKNLI HTTATNLATS ELGSDINDEG DNIFNGSLIR GTFNNNSI F DSKVHKHRTT TYVEGISTVE QLIQKFLDYS STLLNDSLLT REESLRQLRK MLDFIFHFNN YIVQVKKVLV LLNHELFNE YSKEFPTKFE KPMDQESIDK RFANLSDTFL MQYEKFGENL VTFLATIKQV GERENQGLLE LSNRLELCFP E UniProtKB: Spindle pole body component |
-Macromolecule #3: Spindle pole body component
| Macromolecule | Name: Spindle pole body component / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candida albicans (yeast) Candida albicans (yeast) |
| Molecular weight | Theoretical: 92.29618 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MALNKVQLIK LYSNRLVKSL VPVEFGEAFI QSIINDLQTT LLNTSSEEQN LSIIINKLKM QFLSNNLKNE WVEFQNIVNS LSKFKSLDQ ICNYLAFLDA LRDEKPEDIL STSTASLSPG KQNLMINTVN TALTLSQLIE PYYDTLSEQT ILTYLPYTML G SDSKIFTF ...String: MALNKVQLIK LYSNRLVKSL VPVEFGEAFI QSIINDLQTT LLNTSSEEQN LSIIINKLKM QFLSNNLKNE WVEFQNIVNS LSKFKSLDQ ICNYLAFLDA LRDEKPEDIL STSTASLSPG KQNLMINTVN TALTLSQLIE PYYDTLSEQT ILTYLPYTML G SDSKIFTF SNNYTRLEIP KDINNSFSSL LREVFEFAIL YKQLAIVVDR YKGTLVSAIK TAYIAILEAQ LNKYVNDINN IF NNKPNSI LVVYNSIFPW ISILRFLYRV SNRLNRLDGY EFLTFIYSFT NHGDPKIRGI AVTAFTEVVK PYYNIVEHWI VKG ELIDNN NEFFIIFDQE QNEFNSIIKL LPKKIPAFIK SSDKIFQIGK TLIFLNKYCR ELKWVNQYNV KYSAILFNNH QGLA SMTTN EMIKLIDSQY NEILTFLTQI IQGNNKLFTH VYNFKRFYFM ETNDFIDAIM VKGKDVFNES SVNISSTYLR KVLQD AIQI SSVKNFEYVD RLDSRVLNPQ HGNLGWESFT IEYKIDDLPM SYLFEGHQHL QYLKMFHFLW KLRQLNNLLN WHFEMF NEL NHNVVTKLSS RNRRPLAKSL SIITSIRFHF TQFLNELIAY LSYDVIEENF QQHIVRKLFY NKNDQDLLLN KSFMNLS EI DPNNDLPKFN VNLLTIDELV ELHGTYIDSI INSSLLNEKL KGNETNISYI DQIFNILQTI FNFINTSQEF YSLVVTFG L LVRSDSNANK IELEQDQEDL EFQLHKIKRK IYKDIYQHDY KRQLNDLKND LNRDYNLKDL SKLL UniProtKB: Spindle pole body component |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: OTHER / Details: Gatan Solarus 950 plasma cleaner | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 75 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Number grids imaged: 2 / Number real images: 1399 / Average electron dose: 2.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 2.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)