+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11678 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
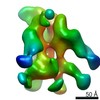
| Title | Cadherin fit into cryo-ET map | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cadherin / Desmosome / CELL ADHESION | |||||||||
| Function / homology |  Function and homology information Function and homology informationcardiac muscle cell-cardiac muscle cell adhesion / Purkinje myocyte development / positive regulation of protein localization to cell-cell junction / bundle of His cell-Purkinje myocyte adhesion involved in cell communication / cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication / desmosome organization / negative regulation of endothelial cell differentiation / Keratinization / negative regulation of inflammatory response to wounding / desmosome ...cardiac muscle cell-cardiac muscle cell adhesion / Purkinje myocyte development / positive regulation of protein localization to cell-cell junction / bundle of His cell-Purkinje myocyte adhesion involved in cell communication / cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication / desmosome organization / negative regulation of endothelial cell differentiation / Keratinization / negative regulation of inflammatory response to wounding / desmosome / mesenchymal to epithelial transition / Formation of the cornified envelope / cornified envelope / regulation of ventricular cardiac muscle cell action potential / Apoptotic cleavage of cell adhesion proteins / positive regulation of p38MAPK cascade / negative regulation of epithelial to mesenchymal transition / positive regulation of sprouting angiogenesis / homophilic cell-cell adhesion / positive regulation of stem cell population maintenance / regulation of heart rate by cardiac conduction / intercalated disc / lateral plasma membrane / RHOG GTPase cycle / RAC2 GTPase cycle / RAC3 GTPase cycle / maternal process involved in female pregnancy / cell adhesion molecule binding / positive regulation of cell adhesion / response to progesterone / cellular response to starvation / stem cell proliferation / adherens junction / cell-cell adhesion / cell-cell junction / cell junction / cytoplasmic vesicle / cell adhesion / apical plasma membrane / intracellular membrane-bounded organelle / calcium ion binding / negative regulation of apoptotic process / cell surface / endoplasmic reticulum / extracellular exosome / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 26.0 Å | |||||||||
 Authors Authors | Sikora M / Ermel UH | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Desmosome architecture derived from molecular dynamics simulations and cryo-electron tomography. Authors: Mateusz Sikora / Utz H Ermel / Anna Seybold / Michael Kunz / Giulia Calloni / Julian Reitz / R Martin Vabulas / Gerhard Hummer / Achilleas S Frangakis /   Abstract: Desmosomes are cell-cell junctions that link tissue cells experiencing intense mechanical stress. Although the structure of the desmosomal cadherins is known, the desmosome architecture-which is ...Desmosomes are cell-cell junctions that link tissue cells experiencing intense mechanical stress. Although the structure of the desmosomal cadherins is known, the desmosome architecture-which is essential for mediating numerous functions-remains elusive. Here, we recorded cryo-electron tomograms (cryo-ET) in which individual cadherins can be discerned; they appear variable in shape, spacing, and tilt with respect to the membrane. The resulting sub-tomogram average reaches a resolution of ∼26 Å, limited by the inherent flexibility of desmosomes. To address this challenge typical of dynamic biological assemblies, we combine sub-tomogram averaging with atomistic molecular dynamics (MD) simulations. We generate models of possible cadherin arrangements and perform an in silico screening according to biophysical and structural properties extracted from MD simulation trajectories. We find a truss-like arrangement of cadherins that resembles the characteristic footprint seen in the electron micrograph. The resulting model of the desmosomal architecture explains their unique biophysical properties and strength. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11678.map.gz emd_11678.map.gz | 536.3 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11678-v30.xml emd-11678-v30.xml emd-11678.xml emd-11678.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11678_fsc.xml emd_11678_fsc.xml | 4.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_11678.png emd_11678.png | 132.1 KB | ||
| Filedesc metadata |  emd-11678.cif.gz emd-11678.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11678 http://ftp.pdbj.org/pub/emdb/structures/EMD-11678 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11678 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11678 | HTTPS FTP |
-Related structure data
| Related structure data |  7a7dMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11678.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11678.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Desmosome from mouse liver
| Entire | Name: Desmosome from mouse liver |
|---|---|
| Components |
|
-Supramolecule #1: Desmosome from mouse liver
| Supramolecule | Name: Desmosome from mouse liver / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Desmoglein-2
| Macromolecule | Name: Desmoglein-2 / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 61.893297 KDa |
| Sequence | String: AWITAPVALR EGEDLSKKNP IAKIHSDLAE ERGLKITYKY TGKGITEPPF GIFVFNKDTG ELNVTSILDR EETPFFLLTG YALDARGNN VEKPLELRIK VLDINDNEPV FTQDVFVGSV EELSAAHTLV MKINATDADE PNTLNSKISY RIVSLEPAYP P VFYLNKDT ...String: AWITAPVALR EGEDLSKKNP IAKIHSDLAE ERGLKITYKY TGKGITEPPF GIFVFNKDTG ELNVTSILDR EETPFFLLTG YALDARGNN VEKPLELRIK VLDINDNEPV FTQDVFVGSV EELSAAHTLV MKINATDADE PNTLNSKISY RIVSLEPAYP P VFYLNKDT GEIYTTSVTL DREEHSSYTL TVEARDGNGE VTDKPVKQAQ VQIRILDVND NIPVVENKVL EGMVEENQVN VE VTRIKVF DADEIGSDNW LANFTFASGN EGGYFHIETD AQTNEGIVTL IKEVDYEEMK NLDFSVIVAN KAAFHKSIRS KYK PTPIPI KVKVKNVKEG IHFKSSVISI YVSESMDRSS KGQIIGNFQA FDEDTGLPAH ARYVKLEDRD NWISVDSVTS EIKL AKLPD FESRYVQNGT YTVKIVAISE DYPRKTITGT VLINVEDIND NCPTLIEPVQ TICHDAEYVN VTAEDLDGHP NSGPF SFSV IDKPPGMAEK WKIARQESTS VLLQQSEKKL GRSEIQFLIS DNQGFSCPEK QVLTLTVCEC LHGSGCREAH UniProtKB: Desmoglein-2 |
-Macromolecule #2: Desmocollin-2
| Macromolecule | Name: Desmocollin-2 / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.86984 KDa |
| Sequence | String: RWAPIPCSML ENSLGPFPLF LQQVQSDTAQ NYTIYYSIRG PGVDQEPRNL FYVERDTGNL YCTRPVDREQ YESFEIIAFA TTPDGYTPE LPLPLIIKIE DENDNYPIFT EETYTFTIFE NCRVGTTVGQ VCATDKDEPD TMHTRLKYSI IGQVPPSPTL F SMHPTTGV ...String: RWAPIPCSML ENSLGPFPLF LQQVQSDTAQ NYTIYYSIRG PGVDQEPRNL FYVERDTGNL YCTRPVDREQ YESFEIIAFA TTPDGYTPE LPLPLIIKIE DENDNYPIFT EETYTFTIFE NCRVGTTVGQ VCATDKDEPD TMHTRLKYSI IGQVPPSPTL F SMHPTTGV ITTTSSQLDR ELIDKYQLKI KVQDMDGQYF GLQTTSTCII NIDDVNDHLP TFTRTSYVTS VEENTVDVEI LR VTVEDKD LVNTANWRAN YTILKGNENG NFKIVTDAKT NEGVLCVVKP LNYEEKQQMI LQIGVVNEAP FSREASPRSA MST ATVTVN VEDQDEGPEC NPPIQTVRMK ENAEVGTTSN GYKAYDPETR SSSGIRYKKL TDPTGWVTID ENTGSIKVFR SLDR EAETI KNGIYNITVL ASDQGGRTCT GTLGIILQDV NDNSPFIPKK TVIICKPTMS SAEIVAVDPD EPIHGPPFDF SLESS TSEV QRMWRLKAIN DTAARLSYQN DPPFGSYVVP ITVRDRLGMS SVTSLDVTLC DCITENDCTH UniProtKB: Desmocollin-2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | tissue |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum SE |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 2.0 sec. / Average electron dose: 1.95 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















