+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11562 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse Hoxa9 IRES like element bound to the human 80S ribosome | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
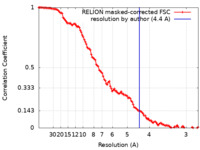
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | ||||||||||||
 Authors Authors | Boehringer D / Lenarcic T / Quade N / Leppek K / Fujii K / Susanto TT / Xue S / Genuth NR / Barna M / Ban N | ||||||||||||
| Funding support |  United States, United States,  Switzerland, 3 items Switzerland, 3 items
| ||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2020 Journal: Mol Cell / Year: 2020Title: Gene- and Species-Specific Hox mRNA Translation by Ribosome Expansion Segments. Authors: Kathrin Leppek / Kotaro Fujii / Nick Quade / Teodorus Theo Susanto / Daniel Boehringer / Tea Lenarčič / Shifeng Xue / Naomi R Genuth / Nenad Ban / Maria Barna /   Abstract: Ribosomes have been suggested to directly control gene regulation, but regulatory roles for ribosomal RNA (rRNA) remain largely unexplored. Expansion segments (ESs) consist of multitudes of tentacle- ...Ribosomes have been suggested to directly control gene regulation, but regulatory roles for ribosomal RNA (rRNA) remain largely unexplored. Expansion segments (ESs) consist of multitudes of tentacle-like rRNA structures extending from the core ribosome in eukaryotes. ESs are remarkably variable in sequence and size across eukaryotic evolution with largely unknown functions. In characterizing ribosome binding to a regulatory element within a Homeobox (Hox) 5' UTR, we identify a modular stem-loop within this element that binds to a single ES, ES9S. Engineering chimeric, "humanized" yeast ribosomes for ES9S reveals that an evolutionary change in the sequence of ES9S endows species-specific binding of Hoxa9 mRNA to the ribosome. Genome editing to site-specifically disrupt the Hoxa9-ES9S interaction demonstrates the functional importance for such selective mRNA-rRNA binding in translation control. Together, these studies unravel unexpected gene regulation directly mediated by rRNA and how ribosome evolution drives translation of critical developmental regulators. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11562.map.gz emd_11562.map.gz | 19.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11562-v30.xml emd-11562-v30.xml emd-11562.xml emd-11562.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11562_fsc.xml emd_11562_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_11562.png emd_11562.png | 165.2 KB | ||
| Masks |  emd_11562_msk_1.map emd_11562_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_11562_half_map_1.map.gz emd_11562_half_map_1.map.gz emd_11562_half_map_2.map.gz emd_11562_half_map_2.map.gz | 98.2 MB 98.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11562 http://ftp.pdbj.org/pub/emdb/structures/EMD-11562 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11562 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11562 | HTTPS FTP |
-Validation report
| Summary document |  emd_11562_validation.pdf.gz emd_11562_validation.pdf.gz | 480.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11562_full_validation.pdf.gz emd_11562_full_validation.pdf.gz | 479.2 KB | Display | |
| Data in XML |  emd_11562_validation.xml.gz emd_11562_validation.xml.gz | 17.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11562 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11562 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11562 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11562 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11562.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11562.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
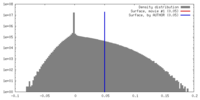
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11562_msk_1.map emd_11562_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
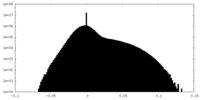
| Density Histograms |
-Half map: #2
| File | emd_11562_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
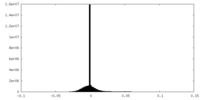
| Density Histograms |
-Half map: #1
| File | emd_11562_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human 80S ribosome - mouse Hoxa9 IRES like element complex
| Entire | Name: human 80S ribosome - mouse Hoxa9 IRES like element complex |
|---|---|
| Components |
|
-Supramolecule #1: human 80S ribosome - mouse Hoxa9 IRES like element complex
| Supramolecule | Name: human 80S ribosome - mouse Hoxa9 IRES like element complex type: complex / ID: 1 / Parent: 0 / Details: Hoxa9 IRES was produced by in vitro transcription |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: HEK293-6E cells Homo sapiens (human) / Strain: HEK293-6E cells |
-Supramolecule #2: 80S ribosome
| Supramolecule | Name: 80S ribosome / type: organelle_or_cellular_component / ID: 2 / Parent: 1 |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)