+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10279 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of SERINC from Drosophila melanogaster | |||||||||
 Map data Map data | main map - auto-sharpened by cryosparc | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Anti-retroviral / TM10 / SERINC fold / novel fold / MEMBRANE PROTEIN | |||||||||
| Function / homology | Serine incorporator/TMS membrane protein / Serine incorporator (Serinc) / membrane / Membrane protein TMS1d Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
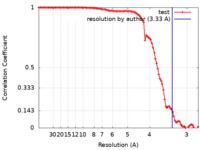
| Method | single particle reconstruction / cryo EM / Resolution: 3.33 Å | |||||||||
 Authors Authors | Pye VE / Nans A | |||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2020 Journal: Nat Struct Mol Biol / Year: 2020Title: A bipartite structural organization defines the SERINC family of HIV-1 restriction factors. Authors: Valerie E Pye / Annachiara Rosa / Cinzia Bertelli / Weston B Struwe / Sarah L Maslen / Robin Corey / Idlir Liko / Mark Hassall / Giada Mattiuzzo / Allison Ballandras-Colas / Andrea Nans / ...Authors: Valerie E Pye / Annachiara Rosa / Cinzia Bertelli / Weston B Struwe / Sarah L Maslen / Robin Corey / Idlir Liko / Mark Hassall / Giada Mattiuzzo / Allison Ballandras-Colas / Andrea Nans / Yasuhiro Takeuchi / Phillip J Stansfeld / J Mark Skehel / Carol V Robinson / Massimo Pizzato / Peter Cherepanov /   Abstract: The human integral membrane protein SERINC5 potently restricts HIV-1 infectivity and sensitizes the virus to antibody-mediated neutralization. Here, using cryo-EM, we determine the structures of ...The human integral membrane protein SERINC5 potently restricts HIV-1 infectivity and sensitizes the virus to antibody-mediated neutralization. Here, using cryo-EM, we determine the structures of human SERINC5 and its orthologue from Drosophila melanogaster at subnanometer and near-atomic resolution, respectively. The structures reveal a novel fold comprised of ten transmembrane helices organized into two subdomains and bisected by a long diagonal helix. A lipid binding groove and clusters of conserved residues highlight potential functional sites. A structure-based mutagenesis scan identified surface-exposed regions and the interface between the subdomains of SERINC5 as critical for HIV-1-restriction activity. The same regions are also important for viral sensitization to neutralizing antibodies, directly linking the antiviral activity of SERINC5 with remodeling of the HIV-1 envelope glycoprotein. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10279.map.gz emd_10279.map.gz | 79 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10279-v30.xml emd-10279-v30.xml emd-10279.xml emd-10279.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10279_fsc.xml emd_10279_fsc.xml | 10.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_10279.png emd_10279.png | 117.4 KB | ||
| Masks |  emd_10279_msk_1.map emd_10279_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10279.cif.gz emd-10279.cif.gz | 6.9 KB | ||
| Others |  emd_10279_additional.map.gz emd_10279_additional.map.gz emd_10279_half_map_1.map.gz emd_10279_half_map_1.map.gz emd_10279_half_map_2.map.gz emd_10279_half_map_2.map.gz | 41.7 MB 77.6 MB 77.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10279 http://ftp.pdbj.org/pub/emdb/structures/EMD-10279 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10279 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10279 | HTTPS FTP |
-Related structure data
| Related structure data |  6sp2MC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10279.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10279.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map - auto-sharpened by cryosparc | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
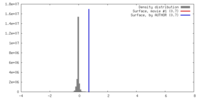
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10279_msk_1.map emd_10279_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
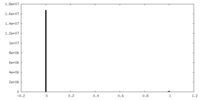
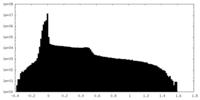
| Density Histograms |
-Additional map: volume map without sharpening
| File | emd_10279_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | volume map without sharpening | ||||||||||||
| Projections & Slices |
| ||||||||||||
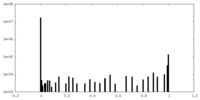
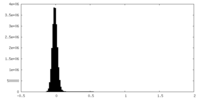
| Density Histograms |
-Half map: half map A
| File | emd_10279_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
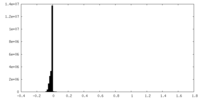
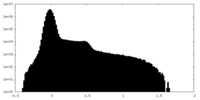
| Density Histograms |
-Half map: half map B
| File | emd_10279_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
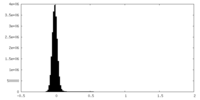
| Density Histograms |
- Sample components
Sample components
-Entire : SERINC homo-hexamer
| Entire | Name: SERINC homo-hexamer |
|---|---|
| Components |
|
-Supramolecule #1: SERINC homo-hexamer
| Supramolecule | Name: SERINC homo-hexamer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: homo-hexamer of SERINC from Drosophila melanogaster recombinantly expressed and purified in detergent micelle; imaged as single particle. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 328.05646 KDa |
-Macromolecule #1: Membrane protein TMS1d
| Macromolecule | Name: Membrane protein TMS1d / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 54.721738 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGAALGICSA AQCAMCCGGT AASMCCSACP SCTNASSSRF MYAFILLVGT VLGAIALSPG LQDTLKKMPF CINSTSSYSS GALSAVSGG SLQVDCEYAL GYMAVYRVCF GMACFFALMS LIMLGVKSSR DPRSHIQNNF WPLKFLICFG AAIGAIFIPD G SFGPAMMW ...String: MGAALGICSA AQCAMCCGGT AASMCCSACP SCTNASSSRF MYAFILLVGT VLGAIALSPG LQDTLKKMPF CINSTSSYSS GALSAVSGG SLQVDCEYAL GYMAVYRVCF GMACFFALMS LIMLGVKSSR DPRSHIQNNF WPLKFLICFG AAIGAIFIPD G SFGPAMMW VGLIGGLAFI LVQLVIIVDF AHSLAENWIE SAENSRGYYY ALAGVTLLCY ILSLTGITLL YIYFTTSTGC GI NKFFISI NLIFCLAISV ISILPAVQER LPHSGLLQSS LVTLYTVYLT WSAVANNPEK ECNPGMFGMM EGFGNATTTA APS THTTRV TFDTTNIIGL VVWLLCILYN CISSAVEVSK ISHDNSEKRV LTEALSDTEA GTDGSGKPST DTETEGVTYS WSMF HLVFV CASLYVMMTL TNWYKPHSEI ELFNGNEASM WVKIVSSWLG VFIYGWSLAA PIVLTNRDFS TGGSSGLEVL FQGPG SGGS AWSHPQFEKG GGSGGGSGGS AWSHPQFEK UniProtKB: Membrane protein TMS1d |
-Macromolecule #2: Lauryl Maltose Neopentyl Glycol
| Macromolecule | Name: Lauryl Maltose Neopentyl Glycol / type: ligand / ID: 2 / Number of copies: 12 / Formula: LMN |
|---|---|
| Molecular weight | Theoretical: 1.005188 KDa |
| Chemical component information |  ChemComp-AV0: |
-Macromolecule #3: O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phospho...
| Macromolecule | Name: O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine type: ligand / ID: 3 / Number of copies: 6 / Formula: P5S |
|---|---|
| Molecular weight | Theoretical: 792.075 Da |
| Chemical component information |  ChemComp-P5S: |
-Macromolecule #4: CARDIOLIPIN
| Macromolecule | Name: CARDIOLIPIN / type: ligand / ID: 4 / Number of copies: 6 / Formula: CDL |
|---|---|
| Molecular weight | Theoretical: 1.464043 KDa |
| Chemical component information |  ChemComp-CDL: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV / Details: wait 60 seconds, blot for 3-4 seconds. | ||||||||||||||||||
| Details | Freshly purified mono-dispersed sample |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average electron dose: 50.0 e/Å2 / Details: 5,807 movies were collected |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 150.6 / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-6sp2: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)