[English] 日本語
 Yorodumi
Yorodumi- EMDB-0638: Structure of the TRPM8 cold receptor by single particle electron ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0638 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the TRPM8 cold receptor by single particle electron cryo-microscopy, TC-I 2014-bound state | |||||||||||||||
 Map data Map data | TRPM8 cold receptor, TC-I 2014-bound state | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | TRANSPORT PROTEIN / Ion Channel / TRPM8 | |||||||||||||||
| Biological species |  Parus major (Great Tit) Parus major (Great Tit) | |||||||||||||||
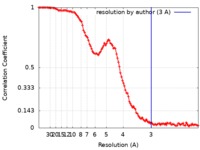
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||||||||
 Authors Authors | Diver MM / Cheng Y / Julius D | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: Structural insights into TRPM8 inhibition and desensitization. Authors: Melinda M Diver / Yifan Cheng / David Julius /  Abstract: The transient receptor potential melastatin 8 (TRPM8) ion channel is the primary detector of environmental cold and an important target for treating pathological cold hypersensitivity. Here, we ...The transient receptor potential melastatin 8 (TRPM8) ion channel is the primary detector of environmental cold and an important target for treating pathological cold hypersensitivity. Here, we present cryo-electron microscopy structures of TRPM8 in ligand-free, antagonist-bound, or calcium-bound forms, revealing how robust conformational changes give rise to two nonconducting states, closed and desensitized. We describe a malleable ligand-binding pocket that accommodates drugs of diverse chemical structures, and we delineate the ion permeation pathway, including the contribution of lipids to pore architecture. Furthermore, we show that direct calcium binding mediates stimulus-evoked desensitization, clarifying this important mechanism of sensory adaptation. We observe large rearrangements within the S4-S5 linker that reposition the S1-S4 and pore domains relative to the TRP helix, leading us to propose a distinct model for modulation of TRPM8 and possibly other TRP channels. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0638.map.gz emd_0638.map.gz | 11 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0638-v30.xml emd-0638-v30.xml emd-0638.xml emd-0638.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0638_fsc.xml emd_0638_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0638.png emd_0638.png | 200.7 KB | ||
| Filedesc metadata |  emd-0638.cif.gz emd-0638.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0638 http://ftp.pdbj.org/pub/emdb/structures/EMD-0638 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0638 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0638 | HTTPS FTP |
-Related structure data
| Related structure data |  6o72MC  0631C  0636C  0639C  6o6aC  6o6rC  6o77C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0638.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0638.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TRPM8 cold receptor, TC-I 2014-bound state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Transient receptor potential cation channel subfamily M member 8
| Entire | Name: Transient receptor potential cation channel subfamily M member 8 |
|---|---|
| Components |
|
-Supramolecule #1: Transient receptor potential cation channel subfamily M member 8
| Supramolecule | Name: Transient receptor potential cation channel subfamily M member 8 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Parus major (Great Tit) Parus major (Great Tit) |
-Macromolecule #1: Transient receptor potential cation channel subfamily M member 8
| Macromolecule | Name: Transient receptor potential cation channel subfamily M member 8 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Parus major (Great Tit) Parus major (Great Tit) |
| Molecular weight | Theoretical: 126.989797 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GAMGSRHRRN GNFESSRLLY SSMSRSIDVA CSDADLANFI QENFKKRECV FFTKDTKSMG NLCKCGYPEN QHIEGTQVNT TEKWNYKKH TKELPTDAFG DIQFENLGKR GKYIRLSCDT DSETLYDLMT QHWHLKTPNL VISVTGGAKN FALKPRMRKI F SRLIYIAQ ...String: GAMGSRHRRN GNFESSRLLY SSMSRSIDVA CSDADLANFI QENFKKRECV FFTKDTKSMG NLCKCGYPEN QHIEGTQVNT TEKWNYKKH TKELPTDAFG DIQFENLGKR GKYIRLSCDT DSETLYDLMT QHWHLKTPNL VISVTGGAKN FALKPRMRKI F SRLIYIAQ SKGAWIFTGG THYGLMKYIG EVVRDNTISR SSEENVVAIG IAAWGMISNR ETLIRTADSD GSFLARYIMD DL KRDPLYC LDNNHTHLLL VDNGTHGHPT TEAKVRTQLE KYISERVIPE SNYGGKIPIV CFAQGGGKET LKSINVAIKS KIP CVVVEG SGRIADVIAS LVEAEGTLAS SCVKESLLRF LPRTISRLSE EETESWIKWI KEVLESPHLL TVIKIEEAGD EIVS NAISF ALYKAFSTNE HDRDNWNGQL KLLLEWNQLD LASDEIFTND RNWESADLQD VMFTALVKDR PKFVRLFLEN GLNLR KFLT TEVLRELYTN NFSSLVFKNL QIAKNSYNDA LLTFVWKMVE DFRRGFKRDY KNSKDEMEIQ LSEECPITRH PLQALF IWS VLQNKKELSK VIWEQTRGCT LAALGASKLL KSMAKVKNDI NAAGESEELA NEYETRAVEL FTECYSNDED LAEQLLT YS CEAWGGSNCL ELAVEARDQQ FIAQPGVQNF LSKQWYGEIS RDTKNWKIIM CLFFFPLIGC GFISFRKKPV EKSKKLFL Y YVSFFTSPFV VFSWNVIFYI AFLLLFAYVL LMDFQKEPTA LEIILYVLVF VLLCDEVRQW YMNGSKYFSD LWNVMDTLA IFYFIAGIVF RLHSDESSWY SGRVIFCLDY IVFTLRLIHI FTVSRNLGPK IIMLQRMMID VFFFLFLFAV WMVAFGVARQ GILRKNEHR WEWIFRSVIY EPYLAMFGQY PDDIDGTTYN FDRCTFSGNE SKPLCVELDA NNQPRFPEWI TIPLVCIYML S TNILLVNL LVAMFGYTVG SVQENNDQVW KFQRFFLVQE YCSRLTIPFP FVIFAYIFMV MRKCFKCCCN KESKEPSICC SR NEDNEIL AWEAVMKENY LVKINTKAND SSEEMVHRFR QLDAKLSDLK GLLKEISSKI K |
-Macromolecule #2: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 2 / Number of copies: 4 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #3: UNDECANE
| Macromolecule | Name: UNDECANE / type: ligand / ID: 3 / Number of copies: 8 / Formula: UND |
|---|---|
| Molecular weight | Theoretical: 156.308 Da |
| Chemical component information | 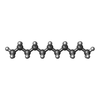 ChemComp-UND: |
-Macromolecule #4: (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoy...
| Macromolecule | Name: (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate type: ligand / ID: 4 / Number of copies: 4 / Formula: 9PE |
|---|---|
| Molecular weight | Theoretical: 593.773 Da |
| Chemical component information | 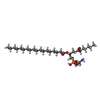 ChemComp-9PE: |
-Macromolecule #5: 3-{7-(trifluoromethyl)-5-[2-(trifluoromethyl)phenyl]-1H-benzimida...
| Macromolecule | Name: 3-{7-(trifluoromethyl)-5-[2-(trifluoromethyl)phenyl]-1H-benzimidazol-2-yl}-1-oxa-2-azaspiro[4.5]dec-2-ene type: ligand / ID: 5 / Number of copies: 4 / Formula: T14 |
|---|---|
| Molecular weight | Theoretical: 467.407 Da |
| Chemical component information | 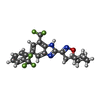 ChemComp-T14: |
-Macromolecule #6: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 6 / Number of copies: 8 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 22500 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)