+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0563 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | LRRC8A-DCPIB in MSP1E3D1 nanodisc expanded state | |||||||||
 Map data Map data | LRRC8A-DCPIB in MSP1E3D1 nanodisc expanded state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion channel / volume-regulation / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMiscellaneous transport and binding events / pre-B cell differentiation / volume-sensitive anion channel activity / aspartate transmembrane transport / cyclic-GMP-AMP transmembrane transporter activity / cyclic-GMP-AMP transmembrane import across plasma membrane / taurine transmembrane transport / monoatomic anion transmembrane transport / protein hexamerization / cell volume homeostasis ...Miscellaneous transport and binding events / pre-B cell differentiation / volume-sensitive anion channel activity / aspartate transmembrane transport / cyclic-GMP-AMP transmembrane transporter activity / cyclic-GMP-AMP transmembrane import across plasma membrane / taurine transmembrane transport / monoatomic anion transmembrane transport / protein hexamerization / cell volume homeostasis / monoatomic anion transport / response to osmotic stress / intracellular glucose homeostasis / monoatomic ion channel complex / positive regulation of myoblast differentiation / chloride transmembrane transport / positive regulation of insulin secretion / spermatogenesis / lysosomal membrane / cell surface / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.32 Å | |||||||||
 Authors Authors | Kern DM / Hite RK | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Cryo-EM structures of the DCPIB-inhibited volume-regulated anion channel LRRC8A in lipid nanodiscs. Authors: David M Kern / SeCheol Oh / Richard K Hite / Stephen G Brohawn /  Abstract: Hypoosmotic conditions activate volume-regulated anion channels in vertebrate cells. These channels are formed by leucine-rich repeat-containing protein 8 (LRRC8) family members and contain LRRC8A in ...Hypoosmotic conditions activate volume-regulated anion channels in vertebrate cells. These channels are formed by leucine-rich repeat-containing protein 8 (LRRC8) family members and contain LRRC8A in homo- or hetero-hexameric assemblies. Here, we present single-particle cryo-electron microscopy structures of LRRC8A in complex with the inhibitor DCPIB reconstituted in lipid nanodiscs. DCPIB plugs the channel like a cork in a bottle - binding in the extracellular selectivity filter and sterically occluding ion conduction. Constricted and expanded structures reveal coupled dilation of cytoplasmic LRRs and the channel pore, suggesting a mechanism for channel gating by internal stimuli. Conformational and symmetry differences between LRRC8A structures determined in detergent micelles and lipid bilayers related to reorganization of intersubunit lipid binding sites demonstrate a critical role for the membrane in determining channel structure. These results provide insight into LRRC8 gating and inhibition and the role of lipids in the structure of an ionic-strength sensing ion channel. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0563.map.gz emd_0563.map.gz | 170.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0563-v30.xml emd-0563-v30.xml emd-0563.xml emd-0563.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0563.png emd_0563.png | 65.8 KB | ||
| Filedesc metadata |  emd-0563.cif.gz emd-0563.cif.gz | 7.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0563 http://ftp.pdbj.org/pub/emdb/structures/EMD-0563 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0563 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0563 | HTTPS FTP |
-Related structure data
| Related structure data |  6nzzMC  0562C  0564C  6nzwC  6o00C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0563.map.gz / Format: CCP4 / Size: 184 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0563.map.gz / Format: CCP4 / Size: 184 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LRRC8A-DCPIB in MSP1E3D1 nanodisc expanded state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.088 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : LRRC8A-DCPIB in MSP1E3D1 nanodisc expanded state
| Entire | Name: LRRC8A-DCPIB in MSP1E3D1 nanodisc expanded state |
|---|---|
| Components |
|
-Supramolecule #1: LRRC8A-DCPIB in MSP1E3D1 nanodisc expanded state
| Supramolecule | Name: LRRC8A-DCPIB in MSP1E3D1 nanodisc expanded state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 565 KDa |
-Macromolecule #1: Volume-regulated anion channel subunit LRRC8A
| Macromolecule | Name: Volume-regulated anion channel subunit LRRC8A / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 95.314562 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIPVTELRYF ADTQPAYRIL KPWWDVFTDY ISIVMLMIAV FGGTLQVTQD KMICLPCKWV TKDSCNDSFR GWAASSPEPT YPNSTVLPT PDTGPTGIKY DLDRHQYNYV DAVCYENRLH WFAKYFPYLV LLHTLIFLAC SNFWFKFPRT SSKLEHFVSI L LKCFDSPW ...String: MIPVTELRYF ADTQPAYRIL KPWWDVFTDY ISIVMLMIAV FGGTLQVTQD KMICLPCKWV TKDSCNDSFR GWAASSPEPT YPNSTVLPT PDTGPTGIKY DLDRHQYNYV DAVCYENRLH WFAKYFPYLV LLHTLIFLAC SNFWFKFPRT SSKLEHFVSI L LKCFDSPW TTRALSETVV EESDPKPAFS KMNGSMDKKS STVSEDVEAT VPMLQRTKSR IEQGIVDRSE TGVLDKKEGE QA KALFEKV KKFRTHVEEG DIVYRLYMRQ TIIKVIKFAL IICYTVYYVH NIKFDVDCTV DIESLTGYRT YRCAHPLATL FKI LASFYI SLVIFYGLIC MYTLWWMLRR SLKKYSFESI REESSYSDIP DVKNDFAFML HLIDQYDPLY SKRFAVFLSE VSEN KLRQL NLNNEWTLDK LRQRLTKNAQ DKLELHLFML SGIPDTVFDL VELEVLKLEL IPDVTIPPSI AQLTGLKELW LYHTA AKIE APALAFLREN LRALHIKFTD IKEIPLWIYS LKTLEELHLT GNLSAENNRY IVIDGLRELK RLKVLRLKSN LSKLPQ VVT DVGVHLQKLS INNEGTKLIV LNSLKKMVNL TELELIRCDL ERIPHSIFSL HNLQEIDLKD NNLKTIEEII SFQHLHR LT CLKLWYNHIA YIPIQIGNLT NLERLYLNRN KIEKIPTQLF YCRKLRYLDL SHNNLTFLPA DIGLLQNLQN LAVTANRI E ALPPELFQCR KLRALHLGNN VLQSLPSRVG ELTNLTQIEL RGNRLECLPV ELGECPLLKR SGLVVEEDLF STLPPEVKE RLWRADKEQA SNSLEVLFQG UniProtKB: Volume-regulated anion channel subunit LRRC8A |
-Macromolecule #2: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 2 / Number of copies: 18 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #3: 4-{[(2S)-2-butyl-6,7-dichloro-2-cyclopentyl-1-oxo-2,3-dihydro-1H-...
| Macromolecule | Name: 4-{[(2S)-2-butyl-6,7-dichloro-2-cyclopentyl-1-oxo-2,3-dihydro-1H-inden-5-yl]oxy}butanoic acid type: ligand / ID: 3 / Number of copies: 1 / Formula: L9Y |
|---|---|
| Molecular weight | Theoretical: 427.361 Da |
| Chemical component information | 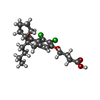 ChemComp-L9Y: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||
| Grid | Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12 / Details: unspecified | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295.15 K / Instrument: FEI VITROBOT MARK IV / Details: 3 microliter drop size, 3 second blot time. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7420 pixel / Digitization - Dimensions - Height: 7676 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 2 / Number real images: 4592 / Average exposure time: 8.0 sec. / Average electron dose: 60.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)





















