[English] 日本語
 Yorodumi
Yorodumi- EMDB-0403: SARS-CoV complex with human neutralizing S230 antibody Fab fragme... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0403 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 1) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | coronavirus spike glycoprotein / MERS-CoV / SARS-CoV / human neutralizing antibodies / Structural Genomics / Seattle Structural Genomics Center for Infectious Disease / SSGCID / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / Attachment and Entry / SARS-CoV-1 activates/modulates innate immune responses / symbiont-mediated-mediated suppression of host tetherin activity / membrane fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell ...Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / Attachment and Entry / SARS-CoV-1 activates/modulates innate immune responses / symbiont-mediated-mediated suppression of host tetherin activity / membrane fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  SARS coronavirus / SARS coronavirus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Walls AC / Xiong X | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2019 Journal: Cell / Year: 2019Title: Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Authors: Alexandra C Walls / Xiaoli Xiong / Young-Jun Park / M Alejandra Tortorici / Joost Snijder / Joel Quispe / Elisabetta Cameroni / Robin Gopal / Mian Dai / Antonio Lanzavecchia / Maria Zambon / ...Authors: Alexandra C Walls / Xiaoli Xiong / Young-Jun Park / M Alejandra Tortorici / Joost Snijder / Joel Quispe / Elisabetta Cameroni / Robin Gopal / Mian Dai / Antonio Lanzavecchia / Maria Zambon / Félix A Rey / Davide Corti / David Veesler /     Abstract: Recent outbreaks of severe acute respiratory syndrome and Middle East respiratory syndrome, along with the threat of a future coronavirus-mediated pandemic, underscore the importance of finding ways ...Recent outbreaks of severe acute respiratory syndrome and Middle East respiratory syndrome, along with the threat of a future coronavirus-mediated pandemic, underscore the importance of finding ways to combat these viruses. The trimeric spike transmembrane glycoprotein S mediates entry into host cells and is the major target of neutralizing antibodies. To understand the humoral immune response elicited upon natural infections with coronaviruses, we structurally characterized the SARS-CoV and MERS-CoV S glycoproteins in complex with neutralizing antibodies isolated from human survivors. Although the two antibodies studied blocked attachment to the host cell receptor, only the anti-SARS-CoV S antibody triggered fusogenic conformational changes via receptor functional mimicry. These results provide a structural framework for understanding coronavirus neutralization by human antibodies and shed light on activation of coronavirus membrane fusion, which takes place through a receptor-driven ratcheting mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0403.map.gz emd_0403.map.gz | 4.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0403-v30.xml emd-0403-v30.xml emd-0403.xml emd-0403.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0403.png emd_0403.png | 37.6 KB | ||
| Filedesc metadata |  emd-0403.cif.gz emd-0403.cif.gz | 6.9 KB | ||
| Others |  emd_0403_additional.map.gz emd_0403_additional.map.gz | 204.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0403 http://ftp.pdbj.org/pub/emdb/structures/EMD-0403 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0403 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0403 | HTTPS FTP |
-Related structure data
| Related structure data |  6nb6MC  0401C  0402C  0404C  6nb3C  6nb4C  6nb5C  6nb7C  6nb8C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0403.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0403.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.37 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
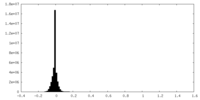
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unsharpened map
| File | emd_0403_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
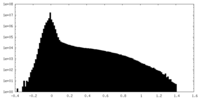
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV S complex with human neutralizing S230 antibody Fab frag...
| Entire | Name: SARS-CoV S complex with human neutralizing S230 antibody Fab fragment (state 1) |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV S complex with human neutralizing S230 antibody Fab frag...
| Supramolecule | Name: SARS-CoV S complex with human neutralizing S230 antibody Fab fragment (state 1) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  SARS coronavirus SARS coronavirus |
| Molecular weight | Theoretical: 572 MDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  SARS coronavirus SARS coronavirus |
| Molecular weight | Theoretical: 139.815969 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GTSDLDRCTT FDDVQAPNYT QHTSSMRGVY YPDEIFRSDT LYLTQDLFLP FYSNVTGFH TINHTFDNPV IPFKDGIYFA ATEKSNVVRG WVFGSTMNNK SQSVIIINNS TNVVIRACNF ELCDNPFFAV S KPMGTQTH ...String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GTSDLDRCTT FDDVQAPNYT QHTSSMRGVY YPDEIFRSDT LYLTQDLFLP FYSNVTGFH TINHTFDNPV IPFKDGIYFA ATEKSNVVRG WVFGSTMNNK SQSVIIINNS TNVVIRACNF ELCDNPFFAV S KPMGTQTH TMIFDNAFNC TFEYISDAFS LDVSEKSGNF KHLREFVFKN KDGFLYVYKG YQPIDVVRDL PSGFNTLKPI FK LPLGINI TNFRAILTAF SPAQDTWGTS AAAYFVGYLK PTTFMLKYDE NGTITDAVDC SQNPLAELKC SVKSFEIDKG IYQ TSNFRV VPSGDVVRFP NITNLCPFGE VFNATKFPSV YAWERKKISN CVADYSVLYN STFFSTFKCY GVSATKLNDL CFSN VYADS FVVKGDDVRQ IAPGQTGVIA DYNYKLPDDF MGCVLAWNTR NIDATSTGNY NYKYRYLRHG KLRPFERDIS NVPFS PDGK PCTPPALNCY WPLNDYGFYT TTGIGYQPYR VVVLSFELLN APATVCGPKL STDLIKNQCV NFNFNGLTGT GVLTPS SKR FQPFQQFGRD VSDFTDSVRD PKTSEILDIS PCSFGGVSVI TPGTNASSEV AVLYQDVNCT DVSTAIHADQ LTPAWRI YS TGNNVFQTQA GCLIGAEHVD TSYECDIPIG AGICASYHTV SLLRSTSQKS IVAYTMSLGA DSSIAYSNNT IAIPTNFS I SITTEVMPVS MAKTSVDCNM YICGDSTECA NLLLQYGSFC TQLNRALSGI AAEQDRNTRE VFAQVKQMYK TPTLKYFGG FNFSQILPDP LKPTKRSFIE DLLFNKVTLA DAGFMKQYGE CLGDINARDL ICAQKFNGLT VLPPLLTDDM IAAYTAALVS GTATAGWTF GAGAALQIPF AMQMAYRFNG IGVTQNVLYE NQKQIANQFN KAISQIQESL TTTSTALGKL QDVVNQNAQA L NTLVKQLS SNFGAISSVL NDILSRLDPP EAEVQIDRLI TGRLQSLQTY VTQQLIRAAE IRASANLAAT KMSECVLGQS KR VDFCGKG YHLMSFPQAA PHGVVFLHVT YVPSQERNFT TAPAICHEGK AYFPREGVFV FNGTSWFITQ RNFFSPQIIT TDN TFVSGN CDVVIGIINN TVYDPLQPEL DSFKEELDKY FKNHTSPDVD LGDISGINAS VVNIQKEIDR LNEVAKNLNE SLID LQELG KYEQYIKGSG RENLYFQGGG GSGYIPEAPR DGQAYVRKDG EWVLLSTFLG HHHHHHHH UniProtKB: Spike glycoprotein |
-Macromolecule #2: S230 heavy chain
| Macromolecule | Name: S230 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.314899 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QAQLVESGGA LVQPGRSLRL SCAASGFTFR NYAMHWVRQA PATGLQWLAV ITSDGRNKFY ADSVKGRFTI SREDSKNTLY LQMDSLRGE DTAVYYCVTQ RDNSRDYFPH YFHDMDVWGQ GTTVAVSS |
-Macromolecule #3: S230 light chain
| Macromolecule | Name: S230 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.329663 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DVVLTQSPLS LPVTLGQPAS ISCRSSQSLV YSDGDTYLNW FQQRPGQSPR RLIYQVSNRD SGVPDRFSGS GSGTDFTLKI SRVEAEDVG VYYCMQGSHW PPTFGQGTKV EIK |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 17 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: PDB 5x58 |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 23071 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION (ver. 3.0) |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-6nb6: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)