+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0292 | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mature MLV capsid hexamer structure in intact virus particles | ||||||||||||||||||||||||||||||
 Map data Map data | None | ||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | MLV / capsid / hexamer / VIRAL PROTEIN | ||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationvirion assembly / viral budding via host ESCRT complex / host multivesicular body / viral nucleocapsid / structural constituent of virion / nucleic acid binding / host cell plasma membrane / RNA binding / zinc ion binding / membrane Similarity search - Function | ||||||||||||||||||||||||||||||
| Biological species |  Murine leukemia virus Murine leukemia virus | ||||||||||||||||||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 7.2 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Qu K / Glass B | ||||||||||||||||||||||||||||||
| Funding support |  Germany, Germany,  United Kingdom, United Kingdom,  Czech Republic, 9 items Czech Republic, 9 items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2018 Journal: Proc Natl Acad Sci U S A / Year: 2018Title: Structure and architecture of immature and mature murine leukemia virus capsids. Authors: Kun Qu / Bärbel Glass / Michal Doležal / Florian K M Schur / Brice Murciano / Alan Rein / Michaela Rumlová / Tomáš Ruml / Hans-Georg Kräusslich / John A G Briggs /      Abstract: Retroviruses assemble and bud from infected cells in an immature form and require proteolytic maturation for infectivity. The CA (capsid) domains of the Gag polyproteins assemble a protein lattice as ...Retroviruses assemble and bud from infected cells in an immature form and require proteolytic maturation for infectivity. The CA (capsid) domains of the Gag polyproteins assemble a protein lattice as a truncated sphere in the immature virion. Proteolytic cleavage of Gag induces dramatic structural rearrangements; a subset of cleaved CA subsequently assembles into the mature core, whose architecture varies among retroviruses. Murine leukemia virus (MLV) is the prototypical γ-retrovirus and serves as the basis of retroviral vectors, but the structure of the MLV CA layer is unknown. Here we have combined X-ray crystallography with cryoelectron tomography to determine the structures of immature and mature MLV CA layers within authentic viral particles. This reveals the structural changes associated with maturation, and, by comparison with HIV-1, uncovers conserved and variable features. In contrast to HIV-1, most MLV CA is used for assembly of the mature core, which adopts variable, multilayered morphologies and does not form a closed structure. Unlike in HIV-1, there is similarity between protein-protein interfaces in the immature MLV CA layer and those in the mature CA layer, and structural maturation of MLV could be achieved through domain rotations that largely maintain hexameric interactions. Nevertheless, the dramatic architectural change on maturation indicates that extensive disassembly and reassembly are required for mature core growth. The core morphology suggests that wrapping of the genome in CA sheets may be sufficient to protect the MLV ribonucleoprotein during cell entry. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0292.map.gz emd_0292.map.gz | 25.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0292-v30.xml emd-0292-v30.xml emd-0292.xml emd-0292.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0292.png emd_0292.png | 446.3 KB | ||
| Filedesc metadata |  emd-0292.cif.gz emd-0292.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0292 http://ftp.pdbj.org/pub/emdb/structures/EMD-0292 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0292 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0292 | HTTPS FTP |
-Related structure data
| Related structure data |  6hwxMC  0290C  0291C  0293C  4419C  4421C  4422C  6gzaC  6hwiC  6hwwC  6hwyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0292.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0292.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
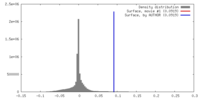
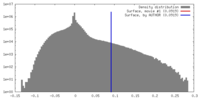
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Murine leukemia virus
| Entire | Name:  Murine leukemia virus Murine leukemia virus |
|---|---|
| Components |
|
-Supramolecule #1: Murine leukemia virus
| Supramolecule | Name: Murine leukemia virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 11786 / Sci species name: Murine leukemia virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: Putative gag polyprotein
| Macromolecule | Name: Putative gag polyprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Murine leukemia virus Murine leukemia virus |
| Molecular weight | Theoretical: 27.165305 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PLRMGGNGQL QYWPFSSSDL YNWKNNNPSF SEDPGKLTAL IESVLTTHQP TWDDCQQLLG TLLTGEEKQR VLLEARKAVR GNDGRPTQL PNEVDAAFPL ERPDWDYTTQ RGRNHLVLYR QLLLAGMQNA GRSPTNLAKV KGITQGPNES PSAFLERLKE A YRRYTPYD ...String: PLRMGGNGQL QYWPFSSSDL YNWKNNNPSF SEDPGKLTAL IESVLTTHQP TWDDCQQLLG TLLTGEEKQR VLLEARKAVR GNDGRPTQL PNEVDAAFPL ERPDWDYTTQ RGRNHLVLYR QLLLAGMQNA GRSPTNLAKV KGITQGPNES PSAFLERLKE A YRRYTPYD PEDPGQETNV SMSFIWQSAP DIGRKLERLE DLKSKTLGDL VREAEKIFNK RETPEEREER IRRETEEK UniProtKB: Putative gag polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Grid | Model: C-flat-2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK II |
| Details | Purified virus solution was inactivated and diluted 1:1 with PBS containing 10 nm colloidal gold. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-6 / Number grids imaged: 2 / Average exposure time: 0.6 sec. / Average electron dose: 1.8 e/Å2 Details: Dose fluctuation was caused by the ring collapse of FEG during data collection. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 7.5 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C6 (6 fold cyclic) / Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 7.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software: (Name: AV3, TOM) / Number subtomograms used: 23397 | ||||||
|---|---|---|---|---|---|---|---|
| Extraction | Number tomograms: 65 / Number images used: 23397 Software:
| ||||||
| Final angle assignment | Type: OTHER / Software: (Name: AV3, TOM) / Details: Subtomogram Averaging. |
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||
| Output model |  PDB-6hwx: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)