[English] 日本語
 Yorodumi
Yorodumi- EMDB-0062: Transient state structure of CRISPR-Cpf1 (Cas12a). I2 conformation -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0062 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Transient state structure of CRISPR-Cpf1 (Cas12a). I2 conformation | |||||||||
 Map data Map data | Transition state CRISPR-Cpf1 (Cas12a)I2 conformation | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR genome editing ribonucleoprotein complex / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationBacillus subtilis ribonuclease / deoxyribonuclease I / deoxyribonuclease I activity / defense response to virus / lyase activity / DNA binding / RNA binding Similarity search - Function | |||||||||
| Biological species |  Francisella tularensis subsp. novicida U112 (bacteria) / Francisella tularensis subsp. novicida U112 (bacteria) /  Francisella tularensis subsp. novicida (strain U112) (bacteria) Francisella tularensis subsp. novicida (strain U112) (bacteria) | |||||||||
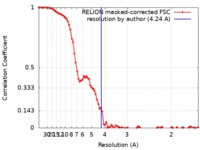
| Method | single particle reconstruction / cryo EM / Resolution: 4.24 Å | |||||||||
 Authors Authors | Montoya G / Mesa P | |||||||||
| Funding support |  Denmark, 1 items Denmark, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2018 Journal: Cell / Year: 2018Title: Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity. Authors: Stella S / Mesa P / Thomsen J / Paul B / Alcon P / Jensen SB / Saligram B / Moses ME / Hatzakis NS / Montoya G | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0062.map.gz emd_0062.map.gz | 34.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0062-v30.xml emd-0062-v30.xml emd-0062.xml emd-0062.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0062_fsc.xml emd_0062_fsc.xml | 8.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_0062.png emd_0062.png | 115 KB | ||
| Filedesc metadata |  emd-0062.cif.gz emd-0062.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0062 http://ftp.pdbj.org/pub/emdb/structures/EMD-0062 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0062 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0062 | HTTPS FTP |
-Validation report
| Summary document |  emd_0062_validation.pdf.gz emd_0062_validation.pdf.gz | 254.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0062_full_validation.pdf.gz emd_0062_full_validation.pdf.gz | 253.6 KB | Display | |
| Data in XML |  emd_0062_validation.xml.gz emd_0062_validation.xml.gz | 10 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0062 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0062 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0062 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0062 | HTTPS FTP |
-Related structure data
| Related structure data |  6gtdMC  0061C  0063C  0064C  0065C  6gtcC  6gteC  6gtfC  6gtgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0062.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0062.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Transition state CRISPR-Cpf1 (Cas12a)I2 conformation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TRansition state complex I2 conformation
| Entire | Name: TRansition state complex I2 conformation |
|---|---|
| Components |
|
-Supramolecule #1: TRansition state complex I2 conformation
| Supramolecule | Name: TRansition state complex I2 conformation / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 180 KDa |
-Supramolecule #2: CRISPR-associated endonuclease Cas12a
| Supramolecule | Name: CRISPR-associated endonuclease Cas12a / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Francisella tularensis subsp. novicida U112 (bacteria) Francisella tularensis subsp. novicida U112 (bacteria) |
-Supramolecule #3: Nucleic acids
| Supramolecule | Name: Nucleic acids / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#4 |
|---|---|
| Source (natural) | Organism:  Francisella tularensis subsp. novicida U112 (bacteria) Francisella tularensis subsp. novicida U112 (bacteria) |
-Macromolecule #1: CRISPR-associated endonuclease Cas12a
| Macromolecule | Name: CRISPR-associated endonuclease Cas12a / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: deoxyribonuclease I |
|---|---|
| Source (natural) | Organism:  Francisella tularensis subsp. novicida (strain U112) (bacteria) Francisella tularensis subsp. novicida (strain U112) (bacteria)Strain: U112 |
| Molecular weight | Theoretical: 155.639828 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSIYQEFVNK YSLSKTLRFE LIPQGKTLEN IKARGLILDD EKRAKDYKKA KQIIDKYHQF FIEEILSSVC ISEDLLQNYS DVYFKLKKS DDDNLQKDFK SAKDTIKKQI SEYIKDSEKF KNLFNQNLID AKKGQESDLI LWLKQSKDNG IELFKANSDI T DIDEALEI ...String: MSIYQEFVNK YSLSKTLRFE LIPQGKTLEN IKARGLILDD EKRAKDYKKA KQIIDKYHQF FIEEILSSVC ISEDLLQNYS DVYFKLKKS DDDNLQKDFK SAKDTIKKQI SEYIKDSEKF KNLFNQNLID AKKGQESDLI LWLKQSKDNG IELFKANSDI T DIDEALEI IKSFKGWTTY FKGFHENRKN VYSSNDIPTS IIYRIVDDNL PKFLENKAKY ESLKDKAPEA INYEQIKKDL AE ELTFDID YKTSEVNQRV FSLDEVFEIA NFNNYLNQSG ITKFNTIIGG KFVNGENTKR KGINEYINLY SQQINDKTLK KYK MSVLFK QILSDTESKS FVIDKLEDDS DVVTTMQSFY EQIAAFKTVE EKSIKETLSL LFDDLKAQKL DLSKIYFKND KSLT DLSQQ VFDDYSVIGT AVLEYITQQI APKNLDNPSK KEQELIAKKT EKAKYLSLET IKLALEEFNK HRDIDKQCRF EEILA NFAA IPMIFDEIAQ NKDNLAQISI KYQNQGKKDL LQASAEDDVK AIKDLLDQTN NLLHKLKIFH ISQSEDKANI LDKDEH FYL VFEECYFELA NIVPLYNKIR NYITQKPYSD EKFKLNFENS TLANGWDKNK EPDNTAILFI KDDKYYLGVM NKKNNKI FD DKAIKENKGE GYKKIVYKLL PGANKMLPKV FFSAKSIKFY NPSEDILRIR NHSTHTKNGS PQKGYEKFEF NIEDCRKF I DFYKQSISKH PEWKDFGFRF SDTQRYNSID EFYREVENQG YKLTFENISE SYIDSVVNQG KLYLFQIYNK DFSAYSKGR PNLHTLYWKA LFDERNLQDV VYKLNGEAEL FYRKQSIPKK ITHPAKEAIA NKNKDNPKKE SVFEYDLIKD KRFTEDKFFF HCPITINFK SSGANKFNDE INLLLKEKAN DVHILSIDRG ERHLAYYTLV DGKGNIIKQD TFNIIGNDRM KTNYHDKLAA I EKDRDSAR KDWKKINNIK EMKEGYLSQV VHEIAKLVIE YNAIVVFQDL NFGFKRGRFK VEKQVYQKLE KMLIEKLNYL VF KDNEFDK TGGVLRAYQL TAPFETFKKM GKQTGIIYYV PAGFTSKICP VTGFVNQLYP KYESVSKSQE FFSKFDKICY NLD KGYFEF SFDYKNFGDK AAKGKWTIAS FGSRLINFRN SDKNHNWDTR EVYPTKELEK LLKDYSIEYG HGECIKAAIC GESD KKFFA KLTSVLNTIL QMRNSKTGTE LDYLISPVAD VNGNFFDSRQ APKNMPQDAD ANGAYHIGLK GLMLLGRIKN NQEGK KLNL VIKNEEYFEF VQNRNNGSEF ELENLYFQGE LRRQASALEH HHHHH UniProtKB: CRISPR-associated endonuclease Cas12a |
-Macromolecule #2: RNA (28-MER)
| Macromolecule | Name: RNA (28-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Francisella tularensis subsp. novicida U112 (bacteria) Francisella tularensis subsp. novicida U112 (bacteria) |
| Molecular weight | Theoretical: 13.749146 KDa |
| Sequence | String: AAUUUCUACU GUUGUAGAUG AGAAGUCAUU UAAUAAGGCC ACU |
-Macromolecule #3: DNA (5'-D(P*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP...
| Macromolecule | Name: DNA (5'-D(P*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP*CP*G)-3') type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Francisella tularensis subsp. novicida U112 (bacteria) Francisella tularensis subsp. novicida U112 (bacteria) |
| Molecular weight | Theoretical: 16.898826 KDa |
| Sequence | String: (DA)(DT)(DT)(DG)(DC)(DT)(DT)(DG)(DC)(DT) (DC)(DG)(DA)(DT)(DG)(DC)(DA)(DT)(DG)(DC) (DA)(DG)(DT)(DG)(DG)(DC)(DC)(DT)(DT) (DA)(DT)(DT)(DA)(DA)(DA)(DT)(DG)(DA)(DC) (DT) (DT)(DC)(DT)(DC)(DT)(DA) ...String: (DA)(DT)(DT)(DG)(DC)(DT)(DT)(DG)(DC)(DT) (DC)(DG)(DA)(DT)(DG)(DC)(DA)(DT)(DG)(DC) (DA)(DG)(DT)(DG)(DG)(DC)(DC)(DT)(DT) (DA)(DT)(DT)(DA)(DA)(DA)(DT)(DG)(DA)(DC) (DT) (DT)(DC)(DT)(DC)(DT)(DA)(DA)(DC) (DG)(DA)(DG)(DC)(DT)(DC)(DG) |
-Macromolecule #4: DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*GP*T)-3')
| Macromolecule | Name: DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*GP*T)-3') type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Francisella tularensis subsp. novicida U112 (bacteria) Francisella tularensis subsp. novicida U112 (bacteria) |
| Molecular weight | Theoretical: 16.992936 KDa |
| Sequence | String: (DC)(DG)(DA)(DG)(DC)(DT)(DC)(DG)(DT)(DT) (DA)(DG)(DA)(DG)(DA)(DA)(DG)(DT)(DC)(DA) (DT)(DT)(DT)(DA)(DA)(DT)(DA)(DA)(DG) (DG)(DC)(DC)(DA)(DC)(DT)(DG)(DC)(DA)(DT) (DG) (DC)(DA)(DT)(DC)(DG)(DA) ...String: (DC)(DG)(DA)(DG)(DC)(DT)(DC)(DG)(DT)(DT) (DA)(DG)(DA)(DG)(DA)(DA)(DG)(DT)(DC)(DA) (DT)(DT)(DT)(DA)(DA)(DT)(DA)(DA)(DG) (DG)(DC)(DC)(DA)(DC)(DT)(DG)(DC)(DA)(DT) (DG) (DC)(DA)(DT)(DC)(DG)(DA)(DG)(DC) (DA)(DA)(DG)(DC)(DA)(DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 4C / Material: COPPER |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)