[English] 日本語
 Yorodumi
Yorodumi- EMDB-0058: Cryo-EM map of a partial yeast 48S preinitiation complex in its c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0058 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of a partial yeast 48S preinitiation complex in its closed conformation obtained by focused classification using masks for eIF3b-eIF3i. | |||||||||
 Map data Map data | For optimal visualization of eIF2, and of eIF3c, eIF3b and eIf3a c-term at the subunit interface, gauss-filter the map by 1.34 and display it at 0.035 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribosome / translation / initiation factors / 40S / eIF1A / eIF3 / eIF2 / tRNAi / 48S PIC / small ribosome subunit | |||||||||
| Function / homology |  Function and homology information Function and homology informationformation of translation initiation ternary complex / Recycling of eIF2:GDP / Cellular response to mitochondrial stress / ABC-family proteins mediated transport / methionyl-initiator methionine tRNA binding / eukaryotic translation initiation factor 3 complex, eIF3e / eukaryotic translation initiation factor 3 complex, eIF3m / incipient cellular bud site / translation reinitiation / eukaryotic translation initiation factor 2 complex ...formation of translation initiation ternary complex / Recycling of eIF2:GDP / Cellular response to mitochondrial stress / ABC-family proteins mediated transport / methionyl-initiator methionine tRNA binding / eukaryotic translation initiation factor 3 complex, eIF3e / eukaryotic translation initiation factor 3 complex, eIF3m / incipient cellular bud site / translation reinitiation / eukaryotic translation initiation factor 2 complex / formation of cytoplasmic translation initiation complex / multi-eIF complex / cytoplasmic translational initiation / eukaryotic translation initiation factor 3 complex / eukaryotic 43S preinitiation complex / formation of translation preinitiation complex / eukaryotic 48S preinitiation complex / positive regulation of translational fidelity / protein-synthesizing GTPase / GDP-dissociation inhibitor activity / Formation of the ternary complex, and subsequently, the 43S complex / Translation initiation complex formation / Ribosomal scanning and start codon recognition / Formation of a pool of free 40S subunits / L13a-mediated translational silencing of Ceruloplasmin expression / ribosomal small subunit binding / 90S preribosome / ribosomal subunit export from nucleus / regulation of translational fidelity / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / translation regulator activity / translation initiation factor binding / translation initiation factor activity / cellular response to amino acid starvation / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / translational initiation / small-subunit processome / maintenance of translational fidelity / cytoplasmic stress granule / rRNA processing / double-stranded RNA binding / regulation of translation / ribosome binding / ribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / cytoplasmic translation / rRNA binding / structural constituent of ribosome / ribosome / translation / G protein-coupled receptor signaling pathway / ribonucleoprotein complex / GTPase activity / mRNA binding / protein kinase binding / GTP binding / nucleolus / RNA binding / zinc ion binding / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |   Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) (yeast) Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) (yeast) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.75 Å | |||||||||
 Authors Authors | Llacer JL / Hussain T | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2015 Journal: Mol Cell / Year: 2015Title: Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex. Authors: Jose L Llácer / Tanweer Hussain / Laura Marler / Colin Echeverría Aitken / Anil Thakur / Jon R Lorsch / Alan G Hinnebusch / V Ramakrishnan /   Abstract: Translation initiation in eukaryotes begins with the formation of a pre-initiation complex (PIC) containing the 40S ribosomal subunit, eIF1, eIF1A, eIF3, ternary complex (eIF2-GTP-Met-tRNAi), and ...Translation initiation in eukaryotes begins with the formation of a pre-initiation complex (PIC) containing the 40S ribosomal subunit, eIF1, eIF1A, eIF3, ternary complex (eIF2-GTP-Met-tRNAi), and eIF5. The PIC, in an open conformation, attaches to the 5' end of the mRNA and scans to locate the start codon, whereupon it closes to arrest scanning. We present single particle cryo-electron microscopy (cryo-EM) reconstructions of 48S PICs from yeast in these open and closed states, at 6.0 Å and 4.9 Å, respectively. These reconstructions show eIF2β as well as a configuration of eIF3 that appears to encircle the 40S, occupying part of the subunit interface. Comparison of the complexes reveals a large conformational change in the 40S head from an open mRNA latch conformation to a closed one that constricts the mRNA entry channel and narrows the P site to enclose tRNAi, thus elucidating key events in start codon recognition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0058.map.gz emd_0058.map.gz | 96.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0058-v30.xml emd-0058-v30.xml emd-0058.xml emd-0058.xml | 83.2 KB 83.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0058.png emd_0058.png | 75.3 KB | ||
| Filedesc metadata |  emd-0058.cif.gz emd-0058.cif.gz | 15.6 KB | ||
| Others |  emd_0058_half_map_1.map.gz emd_0058_half_map_1.map.gz emd_0058_half_map_2.map.gz emd_0058_half_map_2.map.gz | 80.8 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0058 http://ftp.pdbj.org/pub/emdb/structures/EMD-0058 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0058 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0058 | HTTPS FTP |
-Related structure data
| Related structure data |  6gsnMC  0057C  6gsmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0058.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0058.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | For optimal visualization of eIF2, and of eIF3c, eIF3b and eIf3a c-term at the subunit interface, gauss-filter the map by 1.34 and display it at 0.035 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_0058_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
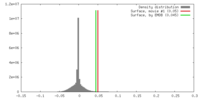
| Density Histograms |
-Half map: #2
| File | emd_0058_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
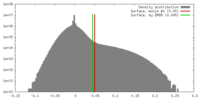
| Density Histograms |
- Sample components
Sample components
+Entire : Structure of a partial yeast 48S preinitiation complex in closed ...
+Supramolecule #1: Structure of a partial yeast 48S preinitiation complex in closed ...
+Supramolecule #3: tRNA
+Supramolecule #4: Initiation factors
+Supramolecule #5: mRNA
+Supramolecule #6: Initiation factors
+Supramolecule #2: Ribosome
+Macromolecule #1: 18S rRNA (1798-MER)
+Macromolecule #20: mRNA
+Macromolecule #28: tRNAi (75-MER)
+Macromolecule #2: KLLA0F09812p
+Macromolecule #3: KLLA0D08305p
+Macromolecule #4: KLLA0D10659p
+Macromolecule #5: KLLA0B08173p
+Macromolecule #6: 40S ribosomal protein S12
+Macromolecule #7: KLLA0F07843p
+Macromolecule #8: 40S ribosomal protein S16
+Macromolecule #9: KLLA0B01474p
+Macromolecule #10: KLLA0B01562p
+Macromolecule #11: KLLA0A07194p
+Macromolecule #12: KLLA0F25542p
+Macromolecule #13: KLLA0B06182p
+Macromolecule #14: 40S ribosomal protein S28
+Macromolecule #15: Ubiquitin-40S ribosomal protein S27a
+Macromolecule #16: KLLA0E12277p
+Macromolecule #17: 40S ribosomal protein S29
+Macromolecule #18: 40S ribosomal protein S30
+Macromolecule #19: 60S ribosomal protein L41-A
+Macromolecule #21: Eukaryotic translation initiation factor 1A
+Macromolecule #22: Eukaryotic translation initiation factor eIF-1
+Macromolecule #23: Eukaryotic translation initiation factor 3 subunit A,eIF3a
+Macromolecule #24: Eukaryotic translation initiation factor 3 subunit C
+Macromolecule #25: Eukaryotic translation initiation factor 2 subunit gamma
+Macromolecule #26: Eukaryotic translation initiation factor 2 subunit beta
+Macromolecule #27: Eukaryotic translation initiation factor 2 subunit alpha
+Macromolecule #29: 40S ribosomal protein S0
+Macromolecule #30: 40S ribosomal protein S1
+Macromolecule #31: 40S ribosomal protein S4
+Macromolecule #32: 40S ribosomal protein S6
+Macromolecule #33: 40S ribosomal protein S7
+Macromolecule #34: 40S ribosomal protein S8
+Macromolecule #35: KLLA0E23673p
+Macromolecule #36: KLLA0A10483p
+Macromolecule #37: KLLA0F18040p
+Macromolecule #38: 40S ribosomal protein S14
+Macromolecule #39: 40S ribosomal protein S21
+Macromolecule #40: 40S ribosomal protein S22
+Macromolecule #41: 40S ribosomal protein S24
+Macromolecule #42: RPS23
+Macromolecule #43: 40S ribosomal protein S26
+Macromolecule #44: 40S ribosomal protein S27
+Macromolecule #45: Eukaryotic translation initiation factor 3 subunit B
+Macromolecule #46: Eukaryotic translation initiation factor 3 subunit I
+Macromolecule #47: Eukaryotic translation initiation factor 3 subunit G
+Macromolecule #48: MAGNESIUM ION
+Macromolecule #49: ZINC ION
+Macromolecule #50: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER
+Macromolecule #51: METHIONINE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.18 mg/mL | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.5 Component:
| ||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 90.0 K / Max: 100.0 K |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 3 / Number real images: 5500 / Average exposure time: 1.1 sec. / Average electron dose: 32.0 e/Å2 Details: Images were collected in movie-mode at 32 frames per second |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Target criteria: FSC |
|---|---|
| Output model |  PDB-6gsn: |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)