+Search query
-Structure paper
| Title | Fragment Screening and Structure-Guided Development of Heparanase Inhibitors Reveal Orthosteric and Allosteric Inhibition |
|---|---|
| Journal, issue, pages | Acs Med. Chem. Lett., Year 2026 |
| Publish date | Apr 3, 2025 (structure data deposition date) |
 Authors Authors | Davies, L.J. / Whitefield, C. / Kim, H. / Nitsche, C. / Jackson, C.J. / Frkic, R.L. |
 External links External links |  Acs Med. Chem. Lett. / Search PubMed Acs Med. Chem. Lett. / Search PubMed |
| Methods | X-ray diffraction |
| Resolution | 1.65 - 2.6 Å |
| Structure data |  PDB-9o1r:  PDB-9o1s:  PDB-9o1t:  PDB-9o1u:  PDB-9o1v:  PDB-9o1w:  PDB-9o1x:  PDB-9o1y:  PDB-9o1z:  PDB-9o20:  PDB-9o21:  PDB-9o22:  PDB-9o23:  PDB-9o24:  PDB-9o25:  PDB-9o26:  PDB-9o27:  PDB-9o28:  PDB-9o29:  PDB-9o2a:  PDB-9o2b:  PDB-9o2c:  PDB-9o2d:  PDB-9o2e:  PDB-9o2f:  PDB-9o2g:  PDB-9o2h:  PDB-9o2i:  PDB-9o2j:  PDB-9o2k:  PDB-9o2l:  PDB-9o2m: |
| Chemicals |  ChemComp-ACT:  ChemComp-PEG:  ChemComp-EDO:  ChemComp-46S:  ChemComp-SO4:  ChemComp-HOH: 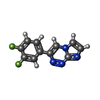 ChemComp-47B:  ChemComp-DMS: 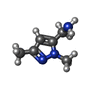 ChemComp-47J: 
ChemComp-AJO:  ChemComp-D86: 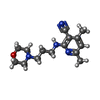 ChemComp-46Y:  ChemComp-PG4: 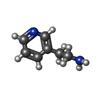 ChemComp-45X:  ChemComp-TP5:  ChemComp-46H: 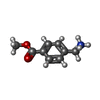 ChemComp-8G2:  PDB-1b72:  ChemComp-4AV:  ChemComp-4AS: 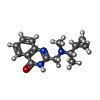 ChemComp-47P: 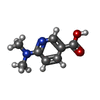 ChemComp-46L:  ChemComp-47Y: 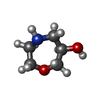 ChemComp-8KB: 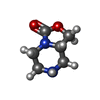 ChemComp-L3Q: 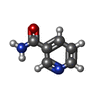 ChemComp-NCA: 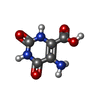 ChemComp-IJZ: 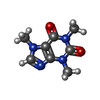 ChemComp-CFF:  ChemComp-GGB:  ChemComp-LNR:  ChemComp-4NC:  ChemComp-3AB:  ChemComp-N7P:  ChemComp-DNC: 
ChemComp-AUM: 
ChemComp-AII:  ChemComp-NVU: 
ChemComp-AQF:  ChemComp-4F8: |
| Source |
|
 Keywords Keywords | HYDROLASE / Heparanase / small molecule / cancer / complex / therapeutics |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 homo sapiens (human)
homo sapiens (human)