+Search query
-Structure paper
| Title | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design. |
|---|---|
| Journal, issue, pages | Cell, Vol. 186, Issue 24, Page 5347-5362.e24, Year 2023 |
| Publish date | Nov 22, 2023 |
 Authors Authors | Pan Shang / Naikang Rong / Jing-Jing Jiang / Jie Cheng / Ming-Hui Zhang / Dongwei Kang / Lei Qi / Lulu Guo / Gong-Ming Yang / Qun Liu / Zhenzhen Zhou / Xiao-Bing Li / Kong-Kai Zhu / Qing-Biao Meng / Xiang Han / Wenqi Yan / Yalei Kong / Lejin Yang / Xiaohui Wang / Dapeng Lei / Xin Feng / Xinyong Liu / Xiao Yu / Yue Wang / Qian Li / Zhen-Hua Shao / Fan Yang / Jin-Peng Sun /  |
| PubMed Abstract | Trace amine-associated receptor 1 (TAAR1) senses a spectrum of endogenous amine-containing metabolites (EAMs) to mediate diverse psychological functions and is useful for schizophrenia treatment ...Trace amine-associated receptor 1 (TAAR1) senses a spectrum of endogenous amine-containing metabolites (EAMs) to mediate diverse psychological functions and is useful for schizophrenia treatment without the side effects of catalepsy. Here, we systematically profiled the signaling properties of TAAR1 activation and present nine structures of TAAR1-Gs/Gq in complex with EAMs, clinical drugs, and synthetic compounds. These structures not only revealed the primary amine recognition pocket (PARP) harboring the conserved acidic D for conserved amine recognition and "twin" toggle switch for receptor activation but also elucidated that targeting specific residues in the second binding pocket (SBP) allowed modulation of signaling preference. In addition to traditional drug-induced Gs signaling, Gq activation by EAM or synthetic compounds is beneficial to schizophrenia treatment. Our results provided a structural and signaling framework for molecular recognition by TAAR1, which afforded structural templates and signal clues for TAAR1-targeted candidate compounds design. |
 External links External links |  Cell / Cell /  PubMed:37963465 PubMed:37963465 |
| Methods | EM (single particle) |
| Resolution | 2.9 - 3.48 Å |
| Structure data | EMDB-37429, PDB-8wc3: EMDB-37430, PDB-8wc4: EMDB-37431, PDB-8wc5: EMDB-37432, PDB-8wc6: EMDB-37433, PDB-8wc7: EMDB-37434, PDB-8wc8: EMDB-37435, PDB-8wc9: EMDB-37436, PDB-8wca: EMDB-37437, PDB-8wcb: EMDB-37438, PDB-8wcc: |
| Chemicals |  ChemComp-UJL:  ChemComp-VMT: 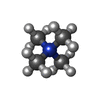 ChemComp-TMA: 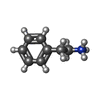 ChemComp-PEA: 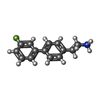 ChemComp-VMK: 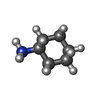 ChemComp-HAI: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / SEP363856 / mTAAR1 / ZH8651 / Gq / TMA / PEA / ZH8667 / hTAAR1 / Gs / CHA |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers























 homo sapiens (human)
homo sapiens (human)
