+Search query
-Structure paper
| Title | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 15, Issue 1, Page 5830, Year 2024 |
| Publish date | Jul 11, 2024 |
 Authors Authors | Hyunji Kang / Ah-Reum Han / Aihua Zhang / Heejin Jeong / Wuhyun Koh / Jung Moo Lee / Hayeon Lee / Hee Young Jo / Miguel A Maria-Solano / Mridula Bhalla / Jea Kwon / Woo Suk Roh / Jimin Yang / Hyun Joo An / Sun Choi / Ho Min Kim / C Justin Lee /  |
| PubMed Abstract | Impaired ion channels regulating Golgi pH lead to structural alterations in the Golgi apparatus, such as fragmentation, which is found, along with cognitive impairment, in Alzheimer's disease. ...Impaired ion channels regulating Golgi pH lead to structural alterations in the Golgi apparatus, such as fragmentation, which is found, along with cognitive impairment, in Alzheimer's disease. However, the causal relationship between altered Golgi structure and cognitive impairment remains elusive due to the lack of understanding of ion channels in the Golgi apparatus of brain cells. Here, we identify that a transmembrane protein TMEM87A, renamed Golgi-pH-regulating cation channel (GolpHCat), expressed in astrocytes and neurons that contributes to hippocampus-dependent memory. We find that GolpHCat displays unique voltage-dependent currents, which is potently inhibited by gluconate. Additionally, we gain structural insights into the ion conduction through GolpHCat at the molecular level by determining three high-resolution cryogenic-electron microscopy structures of human GolpHCat. GolpHCat-knockout mice show fragmented Golgi morphology and altered protein glycosylation and functions in the hippocampus, leading to impaired spatial memory. These findings suggest a molecular target for Golgi-related diseases and cognitive impairment. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:38992057 / PubMed:38992057 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.1 - 3.6 Å |
| Structure data | EMDB-34998, PDB-8hsi: EMDB-35017, PDB-8htt: EMDB-37069, PDB-8kb4: |
| Chemicals |  ChemComp-L9Q:  ChemComp-CLR: 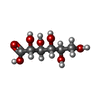 ChemComp-GCO:  ChemComp-NAG: 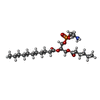 ChemComp-65I: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / non-selective cation channel / ion channel / Golgi-localized protein |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers









 homo sapiens (human)
homo sapiens (human)
 human adenovirus 2
human adenovirus 2