+Search query
-Structure paper
| Title | Promiscuous recognition of MR1 drives self-reactive mucosal-associated invariant T cell responses. |
|---|---|
| Journal, issue, pages | J. Exp. Med., Vol. 220, Year 2023 |
| Publish date | May 9, 2022 (structure data deposition date) |
 Authors Authors | Chancellor, A. / Alan Simmons, R. / Khanolkar, R.C. / Nosi, V. / Beshirova, A. / Berloffa, G. / Colombo, R. / Karuppiah, V. / Pentier, J.M. / Tubb, V. ...Chancellor, A. / Alan Simmons, R. / Khanolkar, R.C. / Nosi, V. / Beshirova, A. / Berloffa, G. / Colombo, R. / Karuppiah, V. / Pentier, J.M. / Tubb, V. / Ghadbane, H. / Suckling, R.J. / Page, K. / Crean, R.M. / Vacchini, A. / De Gregorio, C. / Schaefer, V. / Constantin, D. / Gligoris, T. / Lloyd, A. / Hock, M. / Srikannathasan, V. / Robinson, R.A. / Besra, G.S. / van der Kamp, M.W. / Mori, L. / Calogero, R. / Cole, D.K. / De Libero, G. / Lepore, M. |
 External links External links |  J. Exp. Med. / J. Exp. Med. /  PubMed:37382893 PubMed:37382893 |
| Methods | X-ray diffraction |
| Resolution | 1.84 - 2.4 Å |
| Structure data |  PDB-7zt2:  PDB-7zt3:  PDB-7zt4:  PDB-7zt5:  PDB-7zt7:  PDB-7zt8:  PDB-7zt9: |
| Chemicals |  ChemComp-2LJ:  ChemComp-EDO:  ChemComp-HOH:  ChemComp-JSO: 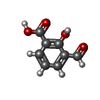 ChemComp-7ZS:  ChemComp-54G: 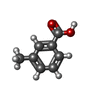 ChemComp-OVV: 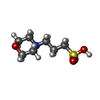 ChemComp-MPO:  ChemComp-4MA: |
| Source |
|
 Keywords Keywords | IMMUNE SYSTEM / TCR / MR1 / 5-OP-RU / 6-Formylpterin / 3-formylsalicylic acid / 5-formylsalicylic acid / 3-formylbenzoic acid / 4-formyl benzoic acid |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 homo sapiens (human)
homo sapiens (human)