+検索条件
-Structure paper
| タイトル | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1. |
|---|---|
| ジャーナル・号・ページ | Nat Chem Biol, Vol. 18, Issue 8, Page 831-840, Year 2022 |
| 掲載日 | 2022年5月30日 |
 著者 著者 | Xin Yang / Xuehui Wang / Zheng Xu / Chao Wu / Yangli Zhou / Yifei Wang / Guifeng Lin / Kan Li / Ming Wu / Anjie Xia / Jingming Liu / Lin Cheng / Jun Zou / Wei Yan / Zhenhua Shao / Shengyong Yang /  |
| PubMed 要旨 | Given the promising clinical value of allosteric modulators of G protein-coupled-receptors (GPCRs), mechanistic understanding of how these modulators alter GPCR function is of significance. Here, we ...Given the promising clinical value of allosteric modulators of G protein-coupled-receptors (GPCRs), mechanistic understanding of how these modulators alter GPCR function is of significance. Here, we report the crystallographic and cryo-electron microscopy structures of the cannabinoid receptor CB1 bound to the positive allosteric modulator (PAM) ZCZ011. These structures show that ZCZ011 binds to an extrahelical site in the transmembrane 2 (TM2)-TM3-TM4 surface. Through (un)biased molecular dynamics simulations and mutagenesis experiments, we show that TM2 rearrangement is critical for the propagation of allosteric signals. ZCZ011 exerts a PAM effect by promoting TM2 rearrangement in favor of receptor activation and increasing the population of receptors that adopt an active conformation. In contrast, ORG27569, a negative allosteric modulator (NAM) of CB1, also binds to the TM2-TM3-TM4 surface and exerts a NAM effect by impeding the TM2 rearrangement. Our findings fill a gap in the understanding of CB1 allosteric regulation and could guide the rational design of CB1 allosteric modulators. |
 リンク リンク |  Nat Chem Biol / Nat Chem Biol /  PubMed:35637350 PubMed:35637350 |
| 手法 | EM (単粒子) / X線回折 |
| 解像度 | 2.7 - 3.36 Å |
| 構造データ | EMDB-32850, PDB-7wv9:  PDB-7fee: |
| 化合物 |  ChemComp-CLR: 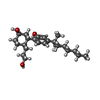 ChemComp-9GF: 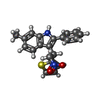 ChemComp-7IC:  ChemComp-PEG:  ChemComp-GOL:  ChemComp-OLC:  ChemComp-OLA:  ChemComp-HOH: |
| 由来 |
|
 キーワード キーワード | MEMBRANE PROTEIN / signal protein / GPCR / Complex / agonist |
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア 万見文献について
万見文献について





 homo sapiens (ヒト)
homo sapiens (ヒト)

 pyrococcus abyssi ge5 (古細菌)
pyrococcus abyssi ge5 (古細菌)