+Search query
-Structure paper
| Title | Cryo-EM structure of human glucose transporter GLUT4. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 13, Issue 1, Page 2671, Year 2022 |
| Publish date | May 13, 2022 |
 Authors Authors | Yafei Yuan / Fang Kong / Hanwen Xu / Angqi Zhu / Nieng Yan / Chuangye Yan /   |
| PubMed Abstract | GLUT4 is the primary glucose transporter in adipose and skeletal muscle tissues. Its cellular trafficking is regulated by insulin signaling. Failed or reduced plasma membrane localization of GLUT4 is ...GLUT4 is the primary glucose transporter in adipose and skeletal muscle tissues. Its cellular trafficking is regulated by insulin signaling. Failed or reduced plasma membrane localization of GLUT4 is associated with diabetes. Here, we report the cryo-EM structures of human GLUT4 bound to a small molecule inhibitor cytochalasin B (CCB) at resolutions of 3.3 Å in both detergent micelles and lipid nanodiscs. CCB-bound GLUT4 exhibits an inward-open conformation. Despite the nearly identical conformation of the transmembrane domain to GLUT1, the cryo-EM structure reveals an extracellular glycosylation site and an intracellular helix that is invisible in the crystal structure of GLUT1. The structural study presented here lays the foundation for further mechanistic investigation of the modulation of GLUT4 trafficking. Our methods for cryo-EM analysis of GLUT4 will also facilitate structural determination of many other small size solute carriers. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:35562357 / PubMed:35562357 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.25 - 3.31 Å |
| Structure data | EMDB-32760, PDB-7wsm: EMDB-32761, PDB-7wsn: |
| Chemicals | 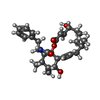 ChemComp-5RH: |
| Source |
|
 Keywords Keywords | TRANSPORT PROTEIN / glucose transporter / GLUT4 / diabetes / cytochalasin B |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers







 homo sapiens (human)
homo sapiens (human)