+Search query
-Structure paper
| Title | Assembly of recombinant tau into filaments identical to those of Alzheimer's disease and chronic traumatic encephalopathy. |
|---|---|
| Journal, issue, pages | Elife, Vol. 11, Year 2022 |
| Publish date | Mar 4, 2022 |
 Authors Authors | Sofia Lövestam / Fujiet Adrian Koh / Bart van Knippenberg / Abhay Kotecha / Alexey G Murzin / Michel Goedert / Sjors H W Scheres /   |
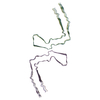
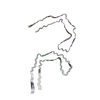
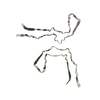
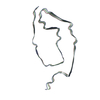
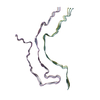
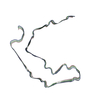
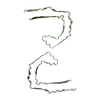
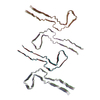
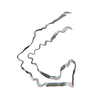
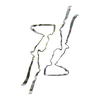
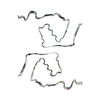
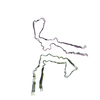
| PubMed Abstract | Abundant filamentous inclusions of tau are characteristic of more than 20 neurodegenerative diseases that are collectively termed tauopathies. Electron cryo-microscopy (cryo-EM) structures of tau ...Abundant filamentous inclusions of tau are characteristic of more than 20 neurodegenerative diseases that are collectively termed tauopathies. Electron cryo-microscopy (cryo-EM) structures of tau amyloid filaments from human brain revealed that distinct tau folds characterise many different diseases. A lack of laboratory-based model systems to generate these structures has hampered efforts to uncover the molecular mechanisms that underlie tauopathies. Here, we report in vitro assembly conditions with recombinant tau that replicate the structures of filaments from both Alzheimer's disease (AD) and chronic traumatic encephalopathy (CTE), as determined by cryo-EM. Our results suggest that post-translational modifications of tau modulate filament assembly, and that previously observed additional densities in AD and CTE filaments may arise from the presence of inorganic salts, like phosphates and sodium chloride. In vitro assembly of tau into disease-relevant filaments will facilitate studies to determine their roles in different diseases, as well as the development of compounds that specifically bind to these structures or prevent their formation. |
 External links External links |  Elife / Elife /  PubMed:35244536 / PubMed:35244536 /  PubMed Central PubMed Central |
| Methods | EM (helical sym.) |
| Resolution | 1.86 - 3.4 Å |
| Structure data | EMDB-14023, PDB-7qjv: EMDB-14024, PDB-7qjw: EMDB-14025, PDB-7qjx: EMDB-14026, PDB-7qjy: EMDB-14027, PDB-7qjz: EMDB-14028, PDB-7qk1: EMDB-14029, PDB-7qk2: EMDB-14030, PDB-7qk3: EMDB-14038, PDB-7qk5: EMDB-14039, PDB-7qk6: EMDB-14040, PDB-7qkf: EMDB-14041, PDB-7qkg: EMDB-14042, PDB-7qkh: EMDB-14043, PDB-7qki: EMDB-14044, PDB-7qkj: EMDB-14045, PDB-7qkk: EMDB-14046, PDB-7qkl: EMDB-14047, PDB-7qkm: EMDB-14053, PDB-7qku: EMDB-14054, PDB-7qkv: EMDB-14055, PDB-7qkw: EMDB-14056, PDB-7qkx: EMDB-14057, PDB-7qky: EMDB-14058, PDB-7qkz: EMDB-14059, PDB-7ql0: EMDB-14060, PDB-7ql1: EMDB-14061, PDB-7ql2: EMDB-14062, PDB-7ql3: EMDB-14063, PDB-7ql4: EMDB-14316, PDB-7r4t: EMDB-14320, PDB-7r5h: |
| Chemicals |  ChemComp-CL:  ChemComp-K: |
| Source |
|
 Keywords Keywords | PROTEIN FIBRIL /  Neurodegeneration / Neurodegeneration /  Amyloid / Tau / Amyloid / Tau /  Alzheimer's Disease / Alzheimer's Disease /  Chronic traumatic encephalopathy Chronic traumatic encephalopathy |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers