+検索条件
-Structure paper
| タイトル | Molecular structures of human ACAT2 disclose mechanism for selective inhibition. |
|---|---|
| ジャーナル・号・ページ | Structure, Vol. 29, Issue 12, Page 1410-11418.e4, Year 2021 |
| 掲載日 | 2021年12月2日 |
 著者 著者 | Tao Long / Yang Liu / Xiaochun Li /  |
| PubMed 要旨 | Endoplasmic reticulum-localized acyl-CoA:cholesterol acyltransferases (ACAT), including ACAT1 and ACAT2, convert cholesterol to cholesteryl esters that become incorporated into lipoproteins or stored ...Endoplasmic reticulum-localized acyl-CoA:cholesterol acyltransferases (ACAT), including ACAT1 and ACAT2, convert cholesterol to cholesteryl esters that become incorporated into lipoproteins or stored in cytosolic lipid droplets. Selective inhibition of ACAT2 has been shown to considerably attenuate hypercholesterolemia and atherosclerosis in mice. Here, we report cryogenic electron microscopy structures of human ACAT2 bound to its specific inhibitor pyripyropene A or the general ACAT inhibitor nevanimibe. Structural analysis reveals that ACAT2 has a topology in membranes similar to that of ACAT1. A catalytic core with an entry site occupied by a cholesterol molecule and another site for allosteric activation of ACAT2 is observed in these structures. Enzymatic assays show that mutations within sites of cholesterol entry or allosteric activation attenuate ACAT2 activity in vitro. Together, these results reveal mechanisms for ACAT2-mediated esterification of cholesterol, providing a blueprint to design new ACAT2 inhibitors for use in the prevention of cardiovascular disease. |
 リンク リンク |  Structure / Structure /  PubMed:34520735 / PubMed:34520735 /  PubMed Central PubMed Central |
| 手法 | EM (単粒子) |
| 解像度 | 3.87 - 3.93 Å |
| 構造データ | EMDB-24208, PDB-7n6q: EMDB-24209, PDB-7n6r: |
| 化合物 | 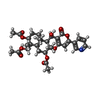 ChemComp-7T8:  ChemComp-CLR:  ChemComp-OLA: 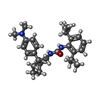 ChemComp-ROV: |
| 由来 |
|
 キーワード キーワード | Transferase/Inhibitor / Inhibitor / MEMBRANE PROTEIN / Transferase-Inhibitor complex |
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア 万見文献について
万見文献について







 homo sapiens (ヒト)
homo sapiens (ヒト)